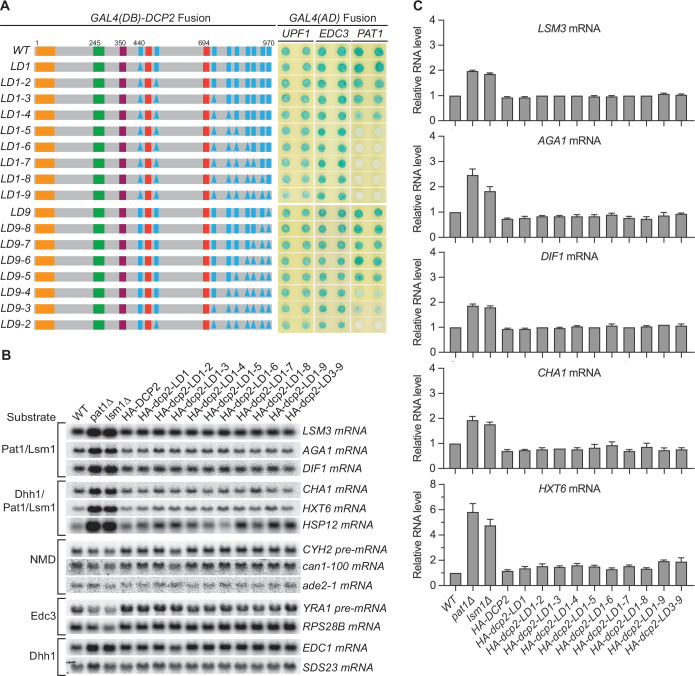
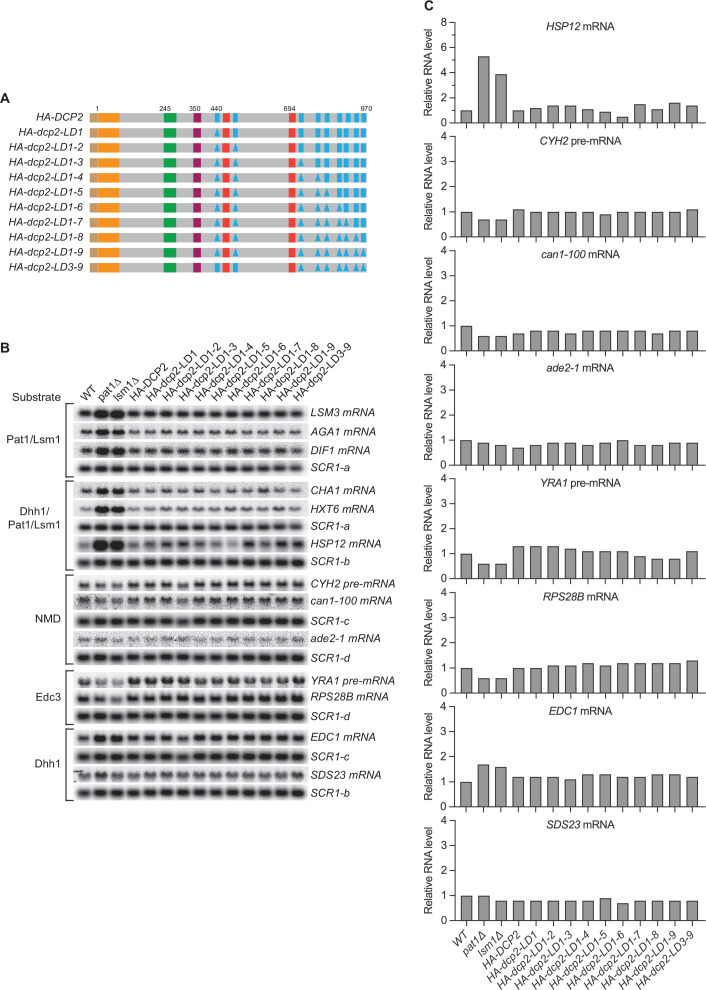
Figure 2. Consequences of deleting Dcp2 leucine-rich Pat1-binding motifs.
(A) Two-hybrid assays evaluating the consequences of deleting the leucine-rich motifs (L1–L9) from the Dcp2 C-terminal domain. As in Figure 1B, schematics of the individual dcp2 alleles are shown on the left (with specific element deletions denoted by triangles) and duplicate two-hybrid assays are on the right. (B) Northern analyses of individual Pat1/Lsm1 and Pat1/Lsm1/Dhh1 substrate levels in cells harboring individual or combined deletions of Dcp2 leucine-rich motifs. (C) Bar graphs of average ± SEM for a subset of the northern analyses depicted in B. The relative levels of each decapping substrate in different strains were determined from three independent experiments, with one representative blot for each transcript shown in panel B. See also Figure 2—figure supplement 1.