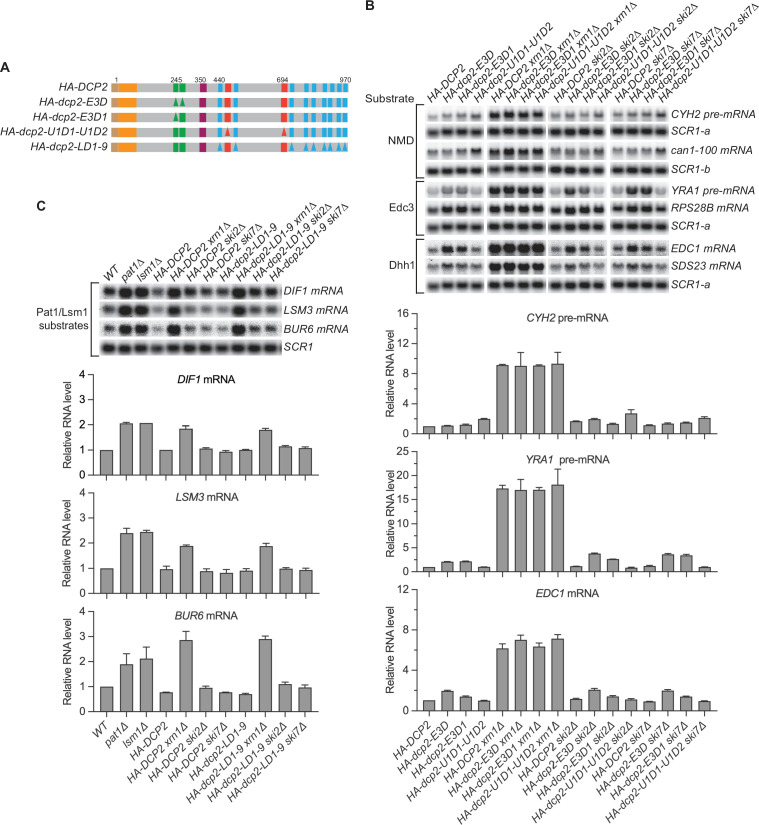
Figure 3. Nonsense-mediated mRNA decay (NMD), Edc3, Dhh1, and Pat1 substrates are still degraded by decapping-dependent 5′ to 3′ decay in the absence of active decapping enzyme recruitment.
(A) Schematics of dcp2 alleles that eliminate Edc3, Upf1, or Pat1 binding to Dcp2. (B) Deletion of XRN1, but not deletions of SKI2 or SKI7, causes significant stabilization of NMD substrates in HA-dcp2-U1D1-U1D2 cells, and Edc3 and Dhh1 substrates in HA-dcp2-E3D or E3D1 cells. (C) Deletion of XRN1, but not deletions of SKI2 or SKI7, causes significant stabilization of Pat1 substrates in HA-dcp2-LD1-9 cells. Northern analyses in B and C as in Figures 1 and 2. Bar graphs in lower panels of B and C depict relative levels of decapping substrates in different strains determined from average ± SEM of three independent experiments. One representative northern blot for each transcript is shown in the upper panels. In the upper panel of B, lower case letters denote SCR1 blots duplicated for clarity of presentation. See also Figure 3—figure supplement 1.
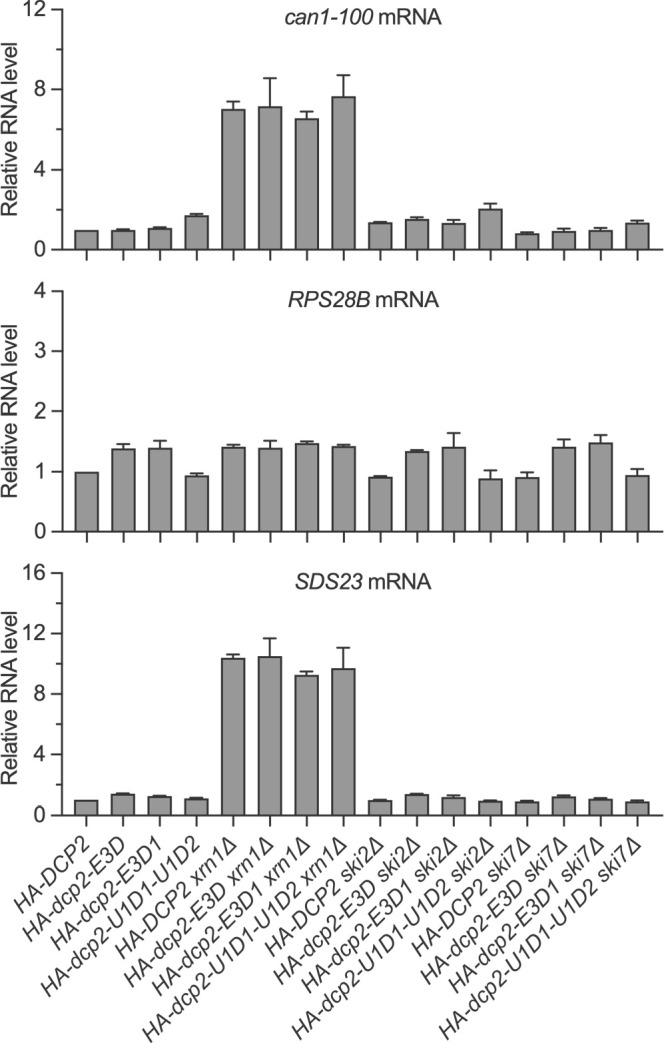
Figure 3—figure supplement 1. Nonsense-mediated mRNA decay (NMD), Edc3, and Dhh1 substrates are still degraded by decapping-dependent 5′ to 3′ decay in the absence of active recruitment of the decapping enzyme.