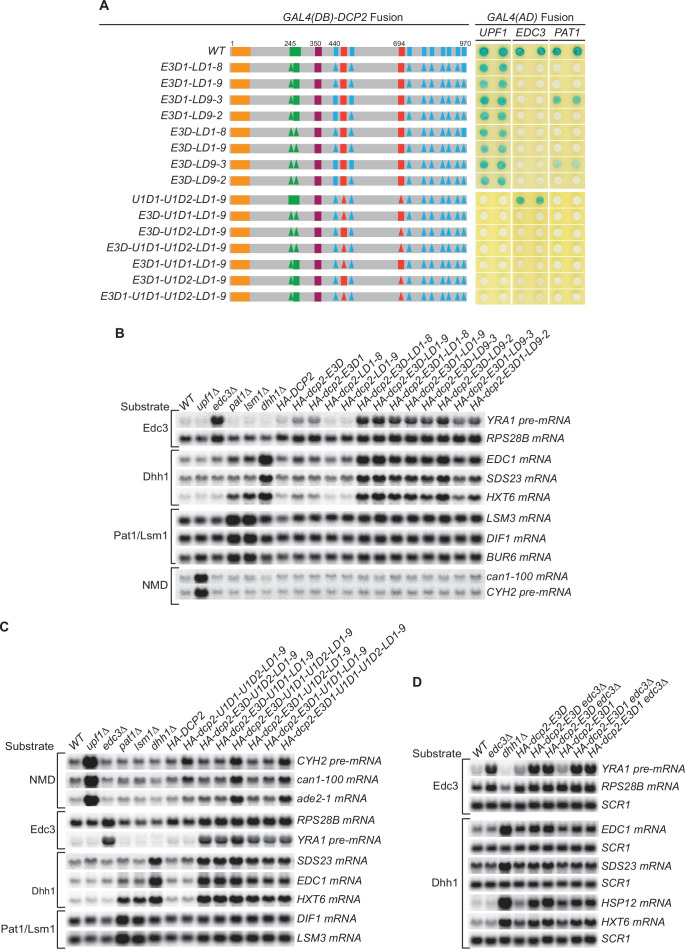
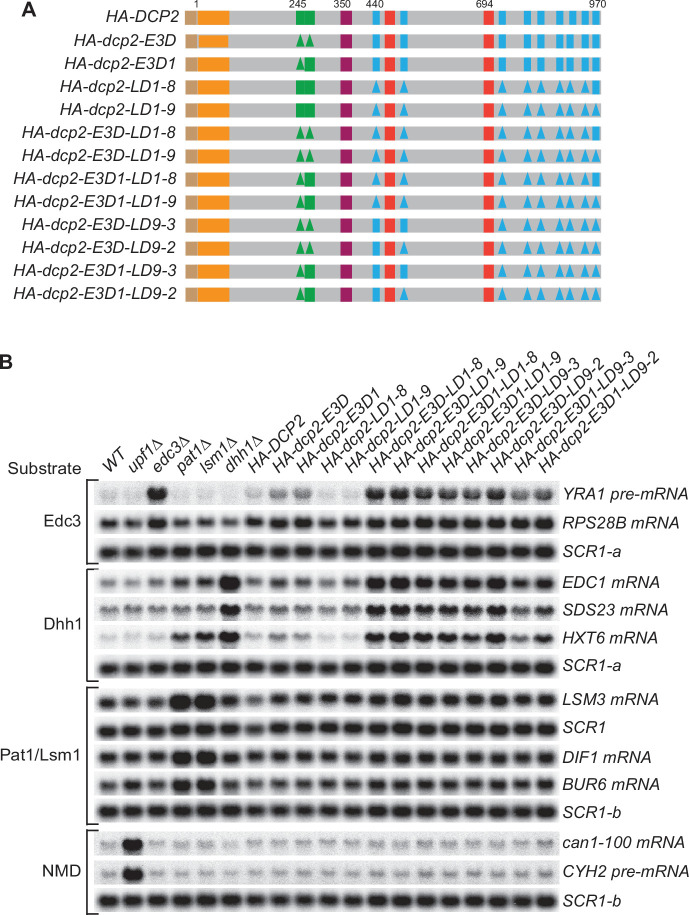
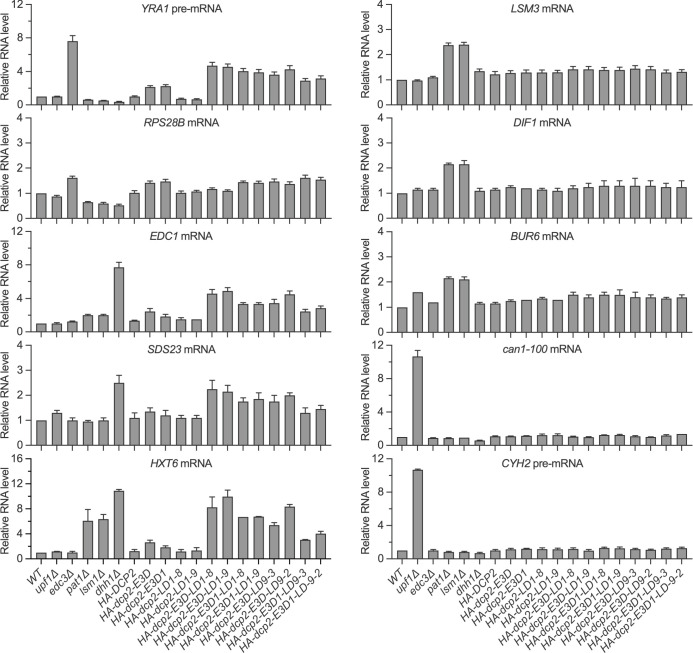
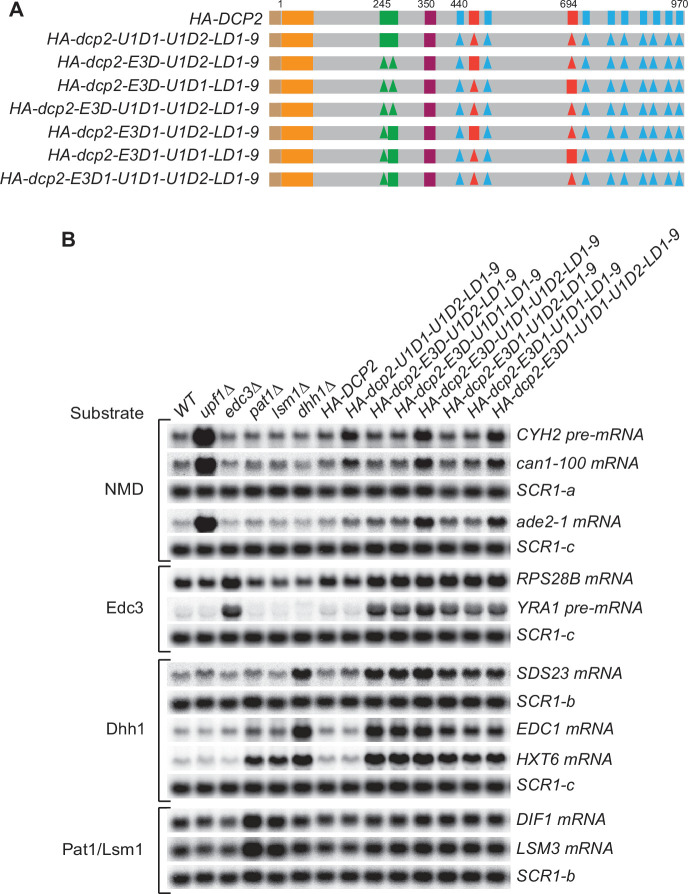
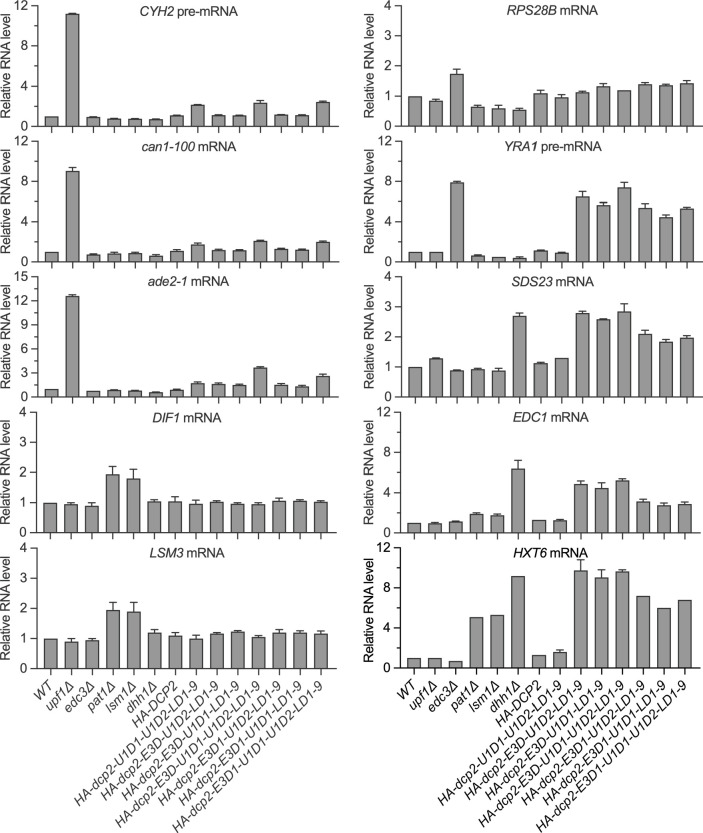
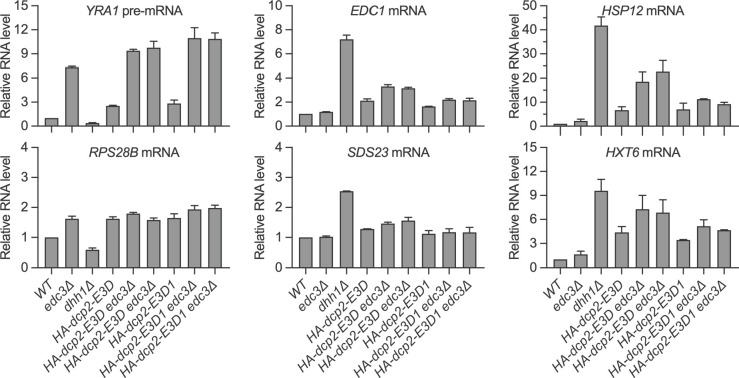
Figure 4. Genetic interactions between the Dcp2 Edc3-, Upf1-, and Pat1-binding motifs, or between the Edc3-binding motif and Edc3, that affect mRNA decapping.
(A) Two-hybrid assays examining the effects of different combinations of element deletions on Edc3, Upf1, and Pat1 binding to Dcp2. Allele schematics and two hybrid analyses are as in Figure 1B. (B) Northern analyses of the consequences of simultaneous loss of the Dcp2 Edc3-binding motif and leucine-rich motifs. See also Figure 4—figure supplement 1 and Figure 4—figure supplement 2. (C) Northern analyses of the consequences of simultaneous loss of the Dcp2 Upf1-binding motifs and leucine-rich motifs, and the roles of single Dcp2 Edc3 or Upf1-binding motifs. See also Figure 4—figure supplement 3 and Figure 4—figure supplement 4. (D) Northern analyses of the consequences for Edc3 and Dhh1 substrates caused by loss of the Dcp2 Edc3-binding motif and trans deletion of EDC3. See also Figure 4—figure supplement 5. Northern analyses as in Figures 1 and 2. Two-hybrid analyses as in Figure 1.