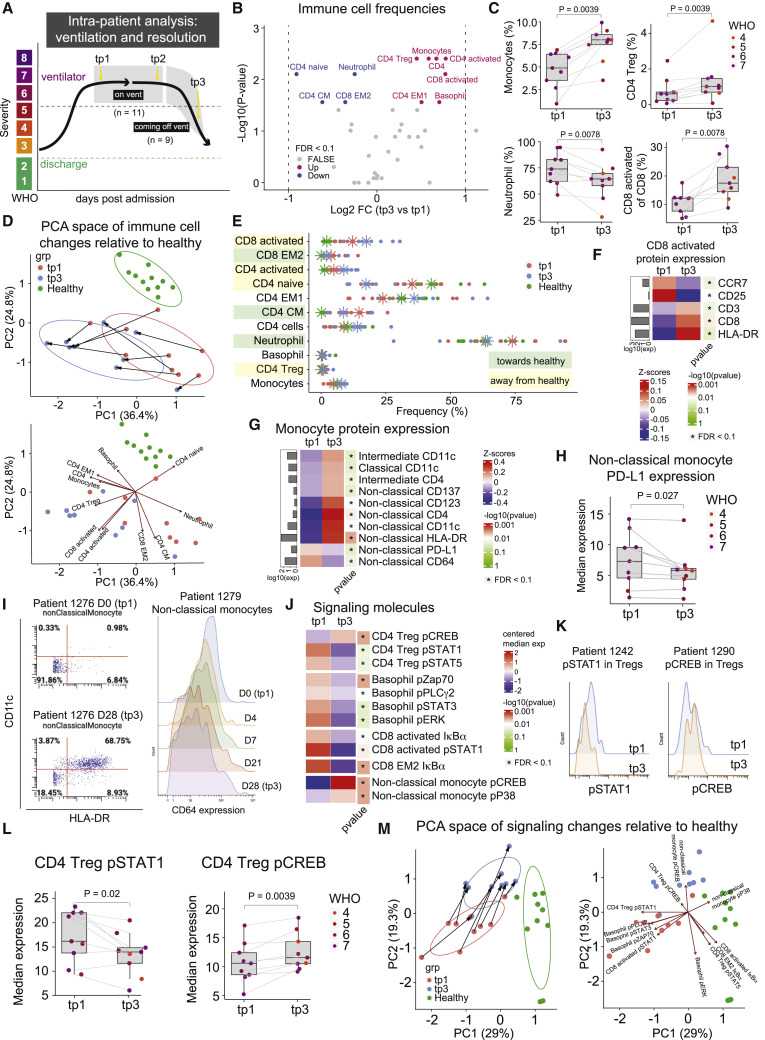
Figure 5.
Recovery from severe COVID-19 requires core immune resolution features and additional regulatory T cell and basophil upregulation
(A) Illustration of intra-patient analysis of ventilated patients. Three timepoints are considered: tp1 (first sample after a patient has been put on a ventilator), tp2 (last sample before the patient is removed from a ventilator), and tp3 (first sample after a patient is successfully removed from ventilation support).
(B) Paired differential abundance analysis of immune cell populations between the first (tp1) and third (tp3) timepoints illustrated in 5A (paired Wilcoxon Rank Sum Test). The log2 fold changes (tp3 versus tp1) are plotted against the negative log10 (nominal p values). Colors indicate whether cell populations are significantly down- (blue) or upregulated (purple) from tp1 to tp3 or not differentially expressed (FALSE, gray) after Benjamini-Hochberg correction, FDR < 0.1.
(C) Frequency of monocytes, neutrophils, CD4 Treg, and CD8 activated T cells at tp1 and tp3. Lines connect samples from the same patient. Nominal p values obtained by paired Wilcoxon Rank Sum Test. CD8 activated T cells are shown as a percentage of parent population (e.g., CD8 T cells), while monocytes, neutrophils, and CD4 Tregs are shown as a percentage of all cells.
(D) Principal component analysis of significant immune cell subsets in 5B for tp1, tp3, and healthy controls. Immune cell directionality and contribution to PCA space denoted on the right.
(E) Population frequencies of significant immune cell subsets in 3B for tp1, tp3, and healthy controls. Stars indicate median value for each group. Cell populations are highlighted in green if tp3 is closer to healthy than tp1 and highlighted in yellow if tp3 is moving away from healthy.
(F and G) Protein expression on CD8 activated T cells (F) and on monocyte subsets (G) at tp1 and tp3. Mean protein expression values have been log10 transformed, scaled, and centered on heatmap. Bars indicate mean protein expression across all samples. Only significant proteins are shown (Wilcoxon Rank Sum Test, Benjamini-Hochberg correction with FDR < 0.1).
(H) Expression of PD-L1 on non-classical monocytes at tp1 and tp3. Lines connect samples from the same patient. Nominal p values obtained by paired Wilcoxon Rank Sum Test.
(I) Left: Scatter plots of CD11c and HLA-DR expression on non-classical monocytes in patient 1276 at day 0 (tp1, top) and day 28 (tp3, bottom). Right: Expression of CD64 on non-classical monocytes for patient 1279 from day 0 (tp1) to day 28 (tp3).
(J) Expression of signaling molecules in significant immune cell subsets in 5B at tp1 and tp3. Median signaling expression values have been centered on heatmap. Only significant signaling molecules are shown (Wilcoxon Rank Sum Test, Benjamini-Hochberg correction with FDR < 0.1 within each cell type).
(K) Expression of pSTAT1 (left) and pCREB (right) in CD4 Tregs at tp1 (blue) and tp3 (orange) for representative patients.
(L) Expression of pSTAT1 and pCREB in CD4 Tregs at tp1 and tp3. Lines connect samples from the same patient. Nominal p values obtained by paired Wilcoxon Rank Sum Test.
(M) Principal component analysis of significant signaling molecules in 5I for tp1, tp3, and healthy controls. Immune cell directionality and contribution to PCA space denoted on the right. See also Figure S5.