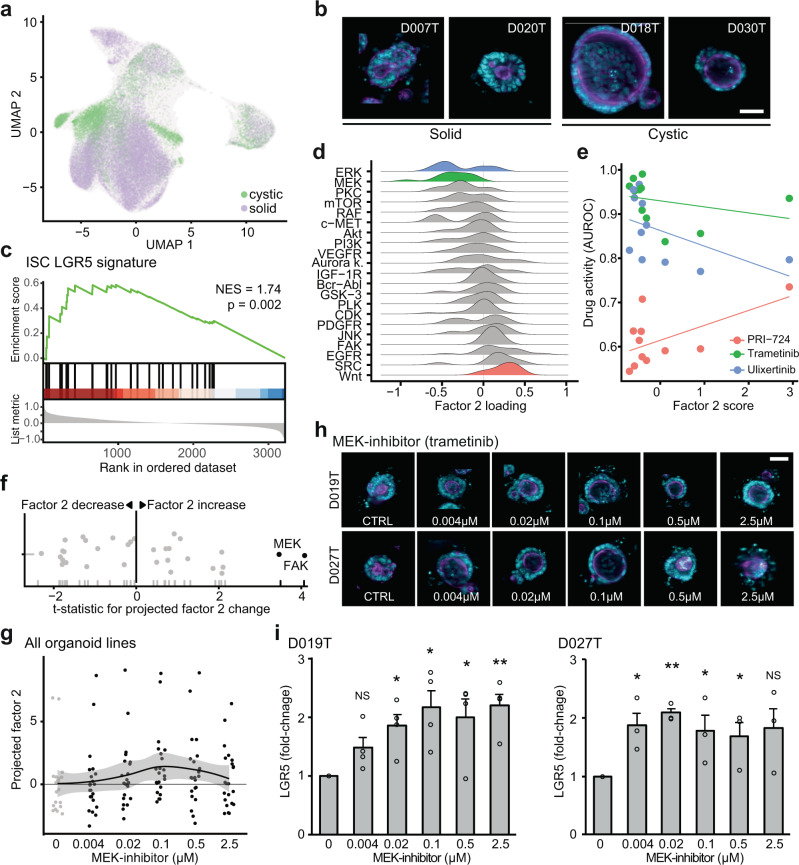
Fig. 5. An LGR5+ stemness program is associated with cystic organoid architecture and can be induced by inhibition of MEK.
a UMAP visualization of cystic and solid organoid morphology in baseline state (DMSO-treated) as defined by factor 2 scores. b Example images of cystic (right) and solid organoid lines. Images were automatically cropped and embedded in black background. Cyan = DNA, magenta = actin; scale bar: 50 µm. c Gene set enrichment analysis of the LGR5+ intestinal stem cell signature34 over ranked factor 2 gene expression loadings (ranking from high factor 2 loading to low factor 2 loading), NES = normalized enrichment score, statistics were calculated with GSEA using 100.000 permutations). d Distributions of drug activity loadings for factor 2 grouped by drug targets. e Relationship of representative drugs’ activity with factor 2 score. Source data are provided as a Source Data file. Further samples can be found in Fig. S7. f Projection of factor 2 scores for drug-induced phenotypes. Highlighted are drug targets leading to a significant change in projected factor scores across all organoid lines (ANOVA). Source data are provided as a Source Data file. g Projected dose-dependent changes in factor 2 scores after treatment with the MEK inhibitor binimetinib across organoid lines. The black line indicates median factor 2 values of 2 replicates from N = 11 organoids treated with binimetinib, grey shading indicates the 95% confidence intervals based on the loess regression with default parameters94. Source data are provided as a Source Data file. h Dose-dependent changes in organoid morphology after treatment with the MEK inhibitor trametinib. Shown are images of organoid lines D019T and D027T (cyan = DNA, magenta = actin; sampled images were cropped and embedded in black background; scale bar: 50 µm) (i), Dose-dependent changes in LGR5 transcript abundance after treatment with the MEK inhibitor trametinib, as assessed by qPCR, data from 3 (D027T) and 4 (D019T) independent replicates are presented as mean + s.e.m. *p < 0.05, **p < 0.005, NS = not significant, two-sided Student’s t test. p values: D019T: p = 0.061 (0.004 µM), p = 0.0196, (0.02 µM), p = 0.0187 (0.1 µM), p = 0.024 (0.5 µM), P = 0.0024 (2.5 µM), D027T: p = 0.0051 (0.004 µM), p = 0.00038, (0.02 µM), p = 0.045 (0.1 µM), p = 0.048 (0.5 µM), P = 0.090 (2.5 µM). Source data are provided as a Source Data file.