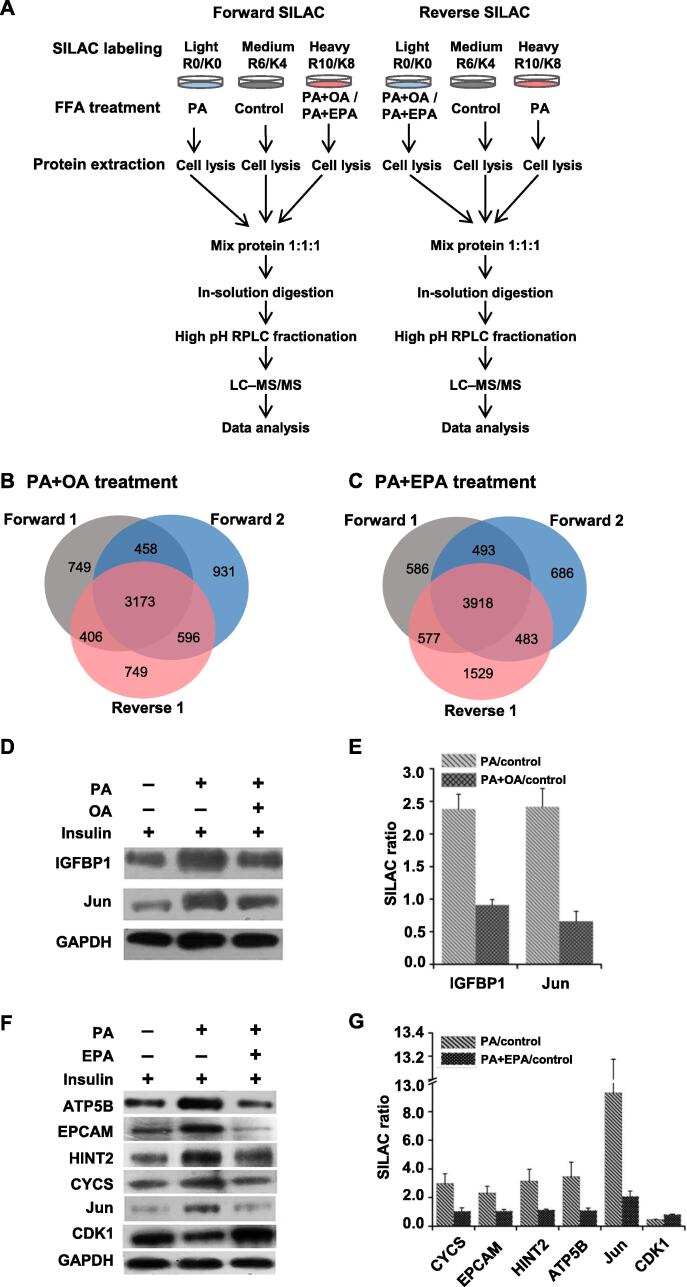
Figure 2.
SILAC-based quantitative proteomic analysis
A. Flow diagram of SILAC labeling combined with LC–MS/MS. B. and C. Quantitation overlap of the triplicate SILAC-based quantitative proteomic experiments upon PA+OA treatment (B) or PA+EPA treatment (C). Forward 1 and Forward 2 represent forward SILAC labeling experiments; Reverse 1 represents reverse SILAC labeling experiment. D. Western blots showing the expression of Jun and IGFBP1 in HepG2 cells treated with PA+OA treatment. E. SILAC quantitative proteomic ratios of Jun and IGFBP1 in HepG2 cells treated with PA+OA. F. Western blots showing the expression of six proteins in HepG2 cells treated with PA+EPA. G. SILAC quantitative proteomic ratios of six proteins in HepG2 cells treated with PA+EPA. SILAC, stable isotope labeling with amino acids in cell culture; RPLC, reversed-phase liquid chromatography; LC–MS/MS, liquid chromatography–tandem mass spectrometry; IGFBP1, insulin-like growth factor-binding protein 1; ATP5B, mitochondrial ATP synthase subunit β; EPCAM, epithelial cell adhesion molecule; HINT2, histidine triad nucleotide-binding protein 2; CYCS, cytochrome c; CDK1, cyclin-dependent kinase 1.