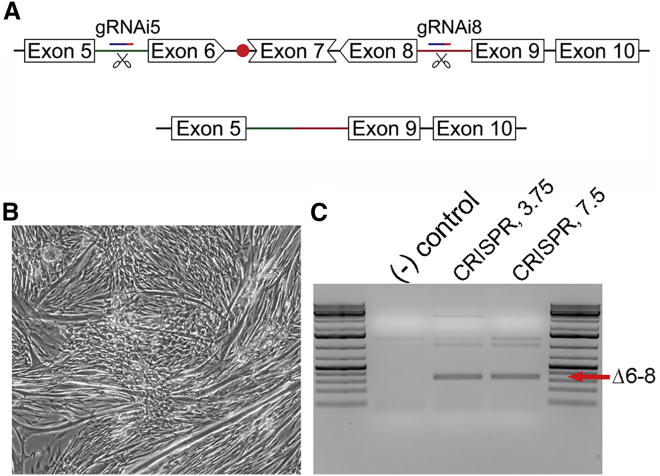
Figure 1.
Generating a dystrophin open reading frame in primary CXMD-derived myoblasts
(A) Outline of the strategy to generate a functional dystrophin open reading frame in CXMD dogs via CRISPR-mediated deletion of exons 6–8, including the splice acceptor mutation in intron 6 (red dot). (B) Primary CXMD-derived myotubes at 5 days post-transfection. (C) Semi-quantitative PCR of DNA isolated from CXMD myotubes at 5 days post-transfection with CRISPR-plasmid using 3.75 and 7.5 μL of lipofectamine. Amplification across the ∼105 kb region targeted for deletion generates a unique Δexons 6–8 deletion product (340 bp; red arrow) in CRISPR-treated samples. The additional products seen likely represent non-specific or incomplete amplification of non-edited genomes due to the large size of the native genomic region being amplified.