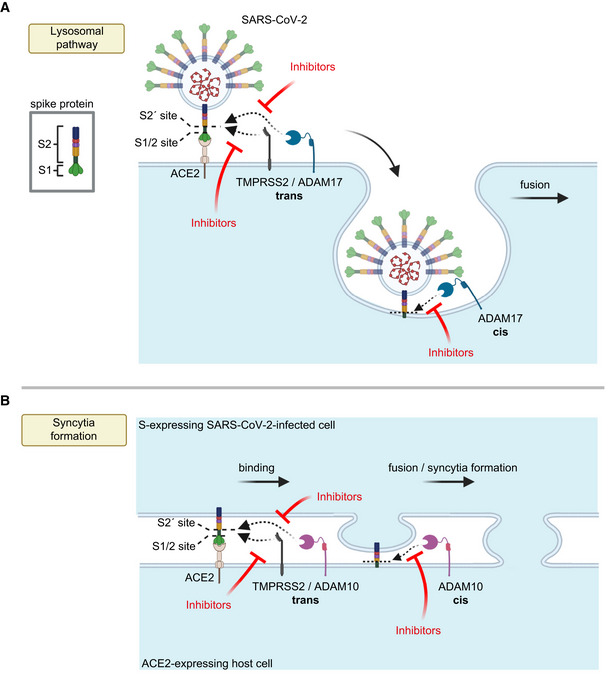
Figure 6. Model for the role of ADAM proteases in SARS‐CoV‐2 viral entry and lung cell syncytia formation.
-
A, BThe SARS‐CoV‐2 spike protein S consists of the two subunits S1 and S2 and binds through S1 to ACE2 on host cells, which is required for viral fusion with host cells (A) and for syncytia formation (B). (A) TMPRSS2 may cleave S in trans and allow for the fusion of SARS‐CoV‐2 with the host cell plasma membrane. In contrast, ADAM17 may either cleave S in trans at the plasma membrane or may cleave it once the fusion peptide of S has inserted into the host plasma membrane after endocytosis, which is conceptually similar to a cleavage in cis. After ADAM17 cleavage, the fusion of SARS‐CoV‐2 with the host cell is likely to happen after endocytosis. Inhibition of ADAM17 reduces viral uptake and fusion. (B) For syncytia formation, the S protein expressed in an infected cell binds at the plasma membrane to ACE2 on another cell. This requires S priming by TMPRSS2 or ADAM10. The priming step may happen in trans, while SARS‐CoV‐2 is still bound to ACE2 or after insertion of the S2 subunit—which contains the fusion peptide—into the membrane of the ACE2‐expressing cell, which may be seen as a cleavage in cis. Inhibitors of TMPRSS2 (e.g., camostat) or ADAM10 reduce cell fusion and syncytia formation. The figure was created with BioRender.
