Abstract
Background
Ralstonia solanacearum causes bacterial wilt of Pogostemon cablin which is an important aromatic herb and also the main materials of COVID-19 therapeutic traditional drugs. However, we are lacking the information on the genomic sequences of R. solanacearum isolated from P. cablin.
Objective
The acquisition and analysis of this whole-genome sequence of the P. cablin bacterial wilt pathogen.
Methods
An R. solanacearum strain, named SY1, was isolated from infected P. cablin plants, and the complete genome sequence was sequenced and analyzed.
Results
The SY1 strain contains a 3.70-Mb chromosome and a 2.18-Mb megaplasmid, with GC contents of 67.57% and 67.41%, respectively. A total of 3308 predicted genes were located on the chromosome and 1657 genes were located in the megaplasmid. SY1 strain has 273 unique genes compared with five representative R. solanacearum strains, and these genes were enriched in the plant–pathogen interaction pathway. SY1 possessed a higher syntenic relationship with phylotype I strains, and the arsenal of type III effectors predicted in SY1 were also more closely related to those of phylotype I strains. SY1 contained 14 and 5 genomic islands in its chromosome and megaplasmid, respectively, and two prophage sequences in its chromosome. In addition, 215 and 130 genes were annotated as carbohydrate-active enzymes and antibiotic resistance genes, respectively.
Conclusion
This is the first genome-scale assembly and annotation for R. solanacearum which isolated from infected P. cablin plants. The arsenal of virulence and antibiotic resistance may as the determinants in SY1 for infection of P. cablin plants.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13258-022-01270-9.
Keywords: Ralstonia solanacearum, Pogostemon cablin, Genome, Effector
Introduction
Ralstonia solanacearum belongs to the Burkholderiaceae (beta-proteobacteria) family and is a gram-negative, soil-born bacterium that causes widespread bacterial wilt in more than 200 plant species in 50 families (Genin and Denny 2012), including many important grain crops and economic crops, such as potato (Solanum tuberosum), tomato (Solanum lycopersicum), banana (Musa sp.), peanut (Arachis hypogaea), ginger (Zingiber officinale), and pepper (Capsicum annuum) (Genin and Denny 2012; Xian et al. 2020). Currently rated as the second most important plant pathogenic bacterium worldwide (Mansfield et al. 2012; Peeters et al. 2013; Chen et al. 2021), R. solanacearum can survive in water and soil for a long time (Álvarez et al. 2008), and invades plants through wounds created on the surface of roots by agricultural activities, infestation of nematodes, or during the formation of lateral roots (Álvarez et al. 2008). Upon penetration into roots, this bacterium gradually invades the xylem vessels, colonizes them, and replicates extensively. The accumulation of bacterial cells and extracellular substances strongly inhibits the ability of the vessel to transport water to the aboveground plant tissues. Finally, the infected plants wilt and die. The bacteria in the dead plants are then returned to the rhizosphere, where they can initiate the next infection cycle (Dalsing et al. 2015).
Phylogenetic analyses of Ralstonia strains that cause wilt diseases revealed great genetic diversity (Genin and Denny 2012; Peeters et al. 2013; Prior et al. 2016). This group of organisms is referred to as the R. solanacearum species complex (RSSC). Members of the RSSC can be divided into four phylotypes according to their geographical origins as follows: phylotype I (Asian), phylotype II (American), phylotype III (African), and phylotype IV (Indonesian) (Peeters et al. 2013). Based on other methods, RSSC members can also be divided into three species as follows: species composed of phylotypes I and III, species composed of phylotype IIA and IIB, and species composed of phylotype IV (Safni et al. 2014; Chen et al. 2021). The virulence factors of R. solanacearum are critical for its pathogenicity (Peeters et al. 2013). The ability to produce extracellular polysaccharides, type IV fimbriae, and polycarboxylate siderophore staphyloferrin B differs between strains in the RSSC (Chen et al. 2021), while the arsenal of type III effector proteins (T3Es) also varies substantially (Peeters et al. 2013; Sun et al. 2017, 2019; Landry et al. 2020).
Pogostemon cablin, commonly called patchouli, is an important aromatic herb belonging to the Lamiaceae family (Li et al. 2020). The essential oil extracted from this plant is an important material in the cosmetics and perfume industries (Phuwajaroanpong et al. 2020). In addition, the plant and its essential oil have been widely used in traditional medicines and pharmacology (Phuwajaroanpong et al. 2020; Aisyah et al. 2021; Junren et al. 2021). P. cablin is native to tropical regions of Asia and is extensively grown in China, India, Brazil, Indonesia, Malaysia, and the Philippines (Wang et al. 2018). P. cablin is susceptible to R. solanacearum infection. The resulting bacterial wilt is difficult to control and causes serious losses in P. cablin (Wang et al., 2018; Zhang et al., 2020).
Since the first report of genome sequencing of the R. solanacearum strain GMI1000, a growing number of other strains have been sequenced (Salanoubat et al. 2002; Chen et al. 2021). There are close to 300 reports of the genome assembly and annotation of R. solanacearum strains in the NCBI database (https://www.ncbi.nlm.nih.gov/genome/490). These data highlight the biodiversity of R. solanacearum strains and have helped to facilitate a better understanding of the evolutionary processes, gene regulatory networks, and pathogenic mechanisms of these bacteria (Prior et al. 2016; Cho et al. 2019). However, genome sequence data of the strain that causes bacterial wilt in P. cablin plants are lacking, and a more comprehensive and detailed analysis of comparative genomics is needed.
In this work, we isolated a R. solanacearum strain, named SY1, from infected P. cablin plants, and sequenced the whole genome of strain SY1. Then, we investigated the host-specific candidate genes, the T3Es, and the evolutionary relationships by comparative evolutionary and genomics analyses.
Materials and methods
Strain Isolation and pathogenicity tests
Ralstonia solanacearum strain SY1 was isolated from the stem leaching solution of one Pogostemon cablin (Blanco) Benth. plant, which exhibited bacterial wilt symptoms, on the Deqing traditional Chinese medicine production base (111°83′ N, 23°25′ E), Deqing City, Guangdong Province of China. The leaching solution of stem were patched on the casamino acid-peptone-glucose (CPG) agar medium for 24 h, as described previously (Tasset et al. 2010). One selected clone was grown in CPG liquid medium for 16 h at 30 °C, followed by centrifugation at 4500 r/min for 5 min to collect cells, and adjusted to 107 CFU/mL in 100 mL of sterile water. Four different cultivars of P. cablin plants (Hainan; Gaoyao; Shipai; Yingni) were cultivated for 60 days (26 ± 2 °C with a 16-h/8-h light/dark photoperiod and 70–80% relative humidity) in the greenhouse, then irrigated with these bacterial suspensions, and the photograph of the plants were taken at 15 days of post-infection.
DNA extraction and genome sequencing and assembly
The genomic DNA of R. solanacearum strain SY1 was extracted using the HiPure Bacterial DNA Kit (Magen Bio, Guangzhou, China) and analyzed for quality via a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and its concentration was determined using a Nanodrop (Thermo Fisher Scientific, Wilmington, USA) following the manufacturer’s instructions. The genomic DNA was sequenced by a PacBio long-read sequencer to obtain the complete genome data. The quality-evaluated genomic DNA was fragmented using g-TUBE (Covaris, Woburn, Massachusetts, USA) followed by end-repair to obtained the SMRTbell libraries. The fragments with sizes larger than 10 kb were selected by the Blue Pippin system according to the manufacturer’s protocols (Pacific Biosciences, Menlo Park, CA, USA). To estimate the quality and average size of the fragments of the library, we used a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) according to standard protocols. SMRT sequencing was done using the PacBio Sequel system (Pacific Biosciences, Menlo Park, CA, USA), and the obtained continuous long reads were used for de novo assembly by the Falcon program (version 0.3.0) with default parameters (Chin et al. 2016). The genome has been deposited in GenBank under accession numbers CP071914 and CP071915 for the chromosome and megaplasmid, respectively.
Functional annotation
The NCBI prokaryotic genome annotation pipeline combined with Prokka (version 1.11) was used to predict the open reading frames (ORFs) (Seemann 2014; Tatusova et al. 2016). CRISPR elements were estimated by CRISPRfinder (version 4.2.17) (Grissa et al. 2007). The rRNAs were predicted using the program rRNAmmer (version 1.2), sRNAs were predicted by the program cmscan (version 1.1.2), and tRNAs were predicted by the program tRNAscan (version 1.3.1) (Lagesen et al. 2007; Nawrocki and Eddy 2013; Lowe and Chan 2016). In addition, tandem repeat elements, interspersed repeat elements, and transposons were predicted by the programs TRF (version 4.09) (Benson 1999), RepeatMasker (version 4.0.5) (Tarailo-Graovac and Chen 2009), and TransposonPSI (version 1.0.0) (http://transposonpsi.sourceforge.net/) (Vij et al. 2016), respectively. The predicted genes of strain SY1 were annotated by BLASTN (E-value < 1e−5) based on sequence similarity, and using GO, Cluster of Orthologous Groups of proteins (COG), KEGG, Swissport, and NCBI nonredundant protein (NR) databases. For protein family annotation, Pfam_Scan (version 1.6) was applied based on the Pfam database (version 32.0) (Finn et al. 2014). GIs and prophages were predicted using Island Viewer (version 4.0) (http://www.pathogenomics.sfu.ca/islandviewer/upload/) (Bertelli et al. 2017) and Phage_Finder (version 2.0) (Fouts 2006), respectively. The type III effectors of the strain SY1 were predicted by the T3E database (Peeters et al. 2013). The phylogenetic tree was generated by the REALPHY program (https://realphy.unibas.ch/realphy/) based on the whole genome of the strain SY1 and other sequenced R. solanacearum genomes (Bertels et al. 2014). The Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al. 2020) and the Carbohydrate-Active enZYmes (CAZy) (Lombard et al. 2013) database were used to perform the advanced annotations. All analysis was conducted using the default parameters.
Identification of orthologous genes
The genome alignments of strain SY1 and five other representative R. solanacearum strains were performed in an all-against-all comparison based on MUMmer 3 package (version 3.3.3) (http://mummer.sourceforge.net/) with default parameters (Kurtz et al. 2004). The orthologous gene clusters in genomes of strains were identified consecutively by combining the programs of DIAMOND (parameters of E-value < 1e–5, query cover > 30%) and OrthoMCL (version 2.0) with default parameters (Buchfink et al. 2015; Silva-Pereira et al. 2019). The putative proteins of strain SY1 and core orthologs were aligned with BLASTP. The score for each pair of significant matched proteins was assigned based on a 1 × 10−7 cut-off value (Hirsh and Fraser 2001). Pairwise comparison of the Average Nucleotide Identity (ANI) was performed by JSpeciesWS (http://jspecies.ribohost.com/jspeciesws/#analyse) (Richter et al. 2015).
Substitution rate estimation
The nonsynonymous mutation rate (Ka) and synonymous mutation rate (Ks) were calculated based on KaKs_Calculator Toolbox software (using the free ratio model with default parameters) (version 2.0) (Wang et al. 2010). The Ka/Ks values higher than 0.5 (Wang et al. 2015) were considered as the positively selected genes within the R. solanacearum strains.
Results
Isolation of pathogenic bacteria and pathogenicity tests
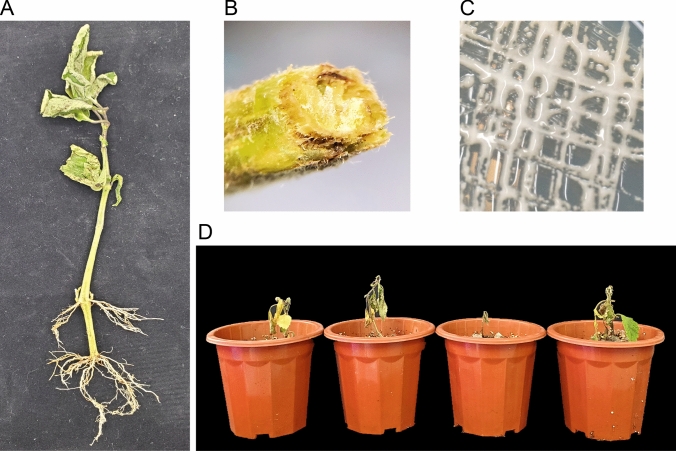
One Pogostemon cablin (Blanco) Benth. plant, which exhibited bacterial wilt symptoms, was sampled from the Deqing traditional Chinese medicine production base (111°83′ N, 23°25′ E), Deqing City, Guangdong Province of China (Fig. 1A). The leaching solution of the stem was cultured and a large number of milky white colonies were obtained (Fig. 1B and C). The back infestation test showed that this strain can caused the typical wilt symptom on four cultivars of P. cablin plants (Hainan; Gaoyao; Shipai; Yingni) which were irrigated with the bacterial suspensions after 15 days of post-infection. Comparison of 16S rRNA gene sequences demonstrated that this strain is a member of the family R. solanacearum, and we named this pathogenic strain SY1.
Fig. 1.
Isolation and pathogenicity tests of Ralstonia solanacearum strain SY1. A and B Ralstonia solanacearum strain SY1 was isolated from the stem of one Pogostemon cabli (Blanco) Benth. plant, which exhibited bacterial wilt symptoms, on the Deqing traditional Chinese medicine production base (111°83′ N, 23°25′ E), Deqing City, Guangdong Province of China. Morphology of R. solanacearum strain SY1 colonies. A single colony of the strain SY1 was streaked and cultivated on CPG medium at 30 °C for 24 h. D Symptoms on P. cablin plants (the cultivars from left to right, Hainan; Gaoyao; Shipai; Yingni) generated by R. solanacearum strain SY1 15 days post-inoculation. Roots of P. cablin plants were irrigated with bacterial suspensions (107 CFU/mL) and photographed 15 days later. CFU, colony‐forming units
Genome sequencing, assembly, and functional annotation
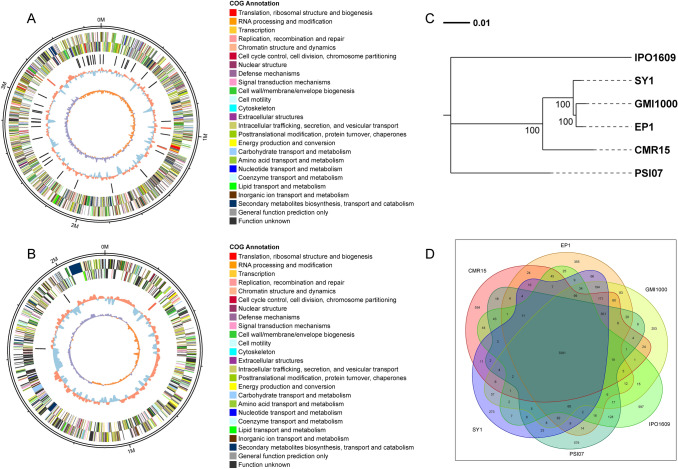
To explore the interaction mechanisms of R. solanacearum strain SY1 with P. cablin, we sequenced the genome of the strain SY1 using the PacBio Sequel platform. A total of 1.50 Gb of polymerase reads with 255-fold coverage of the whole genome were obtained by SMRT sequencing. After removing adapters and low-quality or ambiguous reads, and using the program MECAT (Xiao et al. 2017), we assembled the clean data into two scaffolds, one chromosome and one megaplasmid, of approximately 3.70 Mb and 2.18 Mb, respectively (Fig. 2A and B). A total of 3,308 and 1,657 predicted genes were located in the chromosome and the megaplasmid, respectively. The GC content of the chromosome was 67.57%, and that of the megaplasmid was 67.41% (Table 1). Different strategies were also used to predict the numbers of clustered regularly interspaced short palindromic repeats (CRISPR), genomic islands (GIs), prophages, interspersed repeats, tandem repeats, and transposons in the chromosome and the megaplasmid (Table 1). The Non-Redundant Protein Database (NR) species distribution statistics indicated that 91.74% of the genes belonged to the R. solanacearum, which suggested that the genome we obtained was high quality (Figure S1).
Fig. 2.
General genomic features of Ralstonia solanacearum strain SY1. A Circos plot of the strain SY1 chromosome. The strain SY1 chromosome contig with a full length of 3,702,477 bp, 67.57% GC content, and 3308 predicted genes. B Circos plot of the strain SY1 megaplasmid. The strain SY1 megaplasmid contig with a full length of 2,177,065 bp, 67.41% GC content, and 1657 predicted genes. From the outermost circle to the center indicates the size of the chromosome, the annotated genes (the second and third circles represent the positive and negative strands, respectively, and different colors indicate the different COG functional classifications), non-coding RNAs (black means tRNA and red indicates rRNA), GC content (orange and blue bars indicate that the GC content in this layer is higher or lower than the average GC level of the genome, respectively), and GC skew curve (orange and purple bars indicate that the GC skew curve in this layer is higher or lower than zero, respectively). C Phylogenetic tree constructed based on the whole genome of five representative R. solanacearum strains and the strain SY1. D Venn diagram showing the shared or unique gene families between strain SY1 and the five representative R. solanacearum strains
Table 1.
General feature of the Ralstonia solanacearum strain SY1 genome
| Features | Chromosome | Megaplasmid | Total |
|---|---|---|---|
| Size (bp) | 3702477 | 2177065 | 5879524 |
| G+C content (%) | 67.57 | 67.41 | 67.52 |
| Coding genes | 3308 | 1657 | 4965 |
| tRNA | 55 | 4 | 59 |
| 23S_rRNA | 3 | 1 | 4 |
| 16S_rRNA | 3 | 1 | 4 |
| 5S_rRNA | 3 | 1 | 4 |
| sRNA | 6 | 1 | 7 |
| CRISPR number | 2 | 1 | 3 |
| Genomic islands | 14 | 5 | 19 |
| Prophage | 2 | 0 | 2 |
| Interspersed repeats | 30 | 10 | 40 |
| Tandem repeats | 204 | 119 | 323 |
| Transposon | 1 | 4 | 5 |
Genetic relationship between strain SY1 and other virulent strains
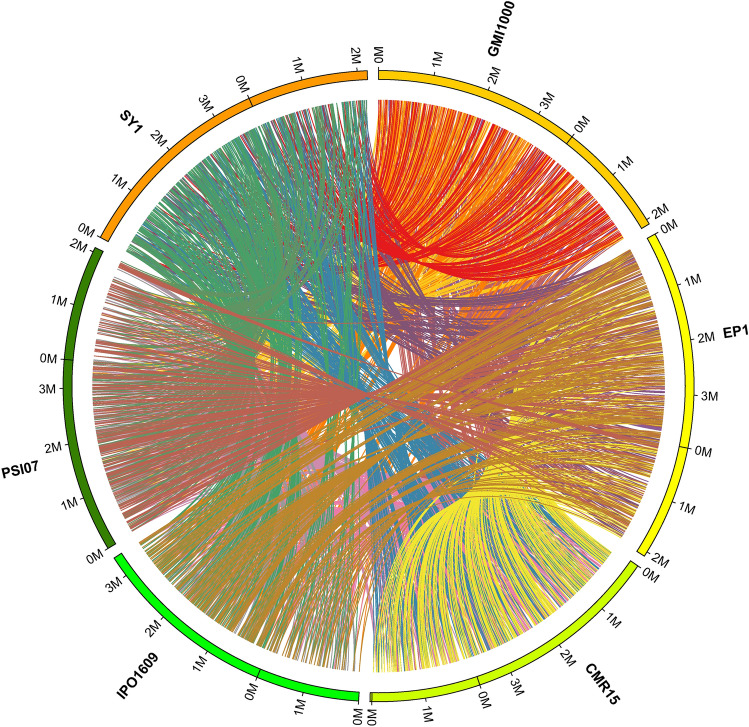
We performed phylogenetic analysis between strain SY1 and five representative sequenced strains of R. solanacearum (GMI1000, EP1, CMR15, IPO1609, and PSI107) based on their whole genomes using the REALPHY platform (Fig. 2C). In addition, in this study, we compared these six R. solanacearum strains and explored the specific and shared genes between the genomes of strain SY1 and these five representative strains (Fig. 2D). We established that 273 genes were unique to strain SY1. Using the C-Sibelia program (Minkin et al., 2013), we identified synteny blocks between strain SY1 and the five representative strains (Fig. 3). In strain SY1, collinear blocks accounted for 92.70% (versus EP1), 91.04% (versus GMI1000), 82.09% (versus CMR15), 76.84% (versus PSI07), and 71.63% (versus IPO1609) of the total gene length (Table 2).
Fig. 3.
Synteny map of the genomes of strain SY1 and five representative R. solanacearum strains: GMI1000, EP1, CMR15, IPO1609, and PSI107
Table 2.
Comparison of collinearity between strain SY1 and other strains of Ralstonia solanacearum
| Strain | Collinear with SY1 (%) |
|---|---|
| EP1 | 92.70 |
| GMI | 1000 91.04 |
| CMR15 | 82.09 |
| PSI07 | 76.84 |
| IPO1609 | 71.63 |
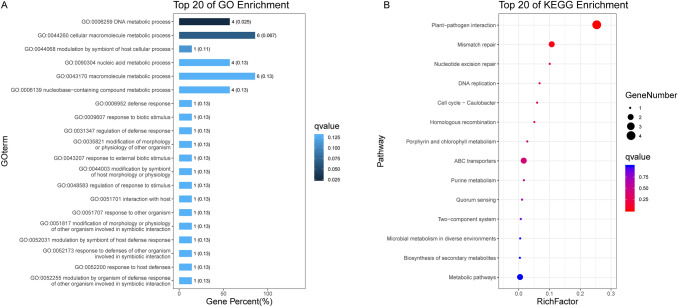
To better evaluate the gene ontology (GO) and functional classification of these strain SY1-specific genes, we performed GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Fig. 4). Genes specifically expressed in strain SY1 were enriched in the GO terms ‘DNA metabolic process’, ‘cellular macromolecule metabolic process’, ‘nucleic acid metabolic process’, and ‘nucleobase-containing compound metabolic process’. Subsequently, we performed KEGG pathway analyses using genes specifically expressed in strain SY1. The main enriched pathways were ‘plant–pathogen interaction’, ‘mismatch repair’, and ‘nucleotide excision repair’ (Table S1 and S2). These results demonstrated that although SY1 has high similarity with other R. solanacearum strains, it still may has many different genes which involve physiology, biochemistry and pathogenicity.
Fig. 4.
Top 20 enriched GO and KEGG pathways of strain SY1-specific genes. The q-value was generated by multi-test correction of the p-value
Identification and comparative analysis of the type III effector of R. solanacearum strain SY1
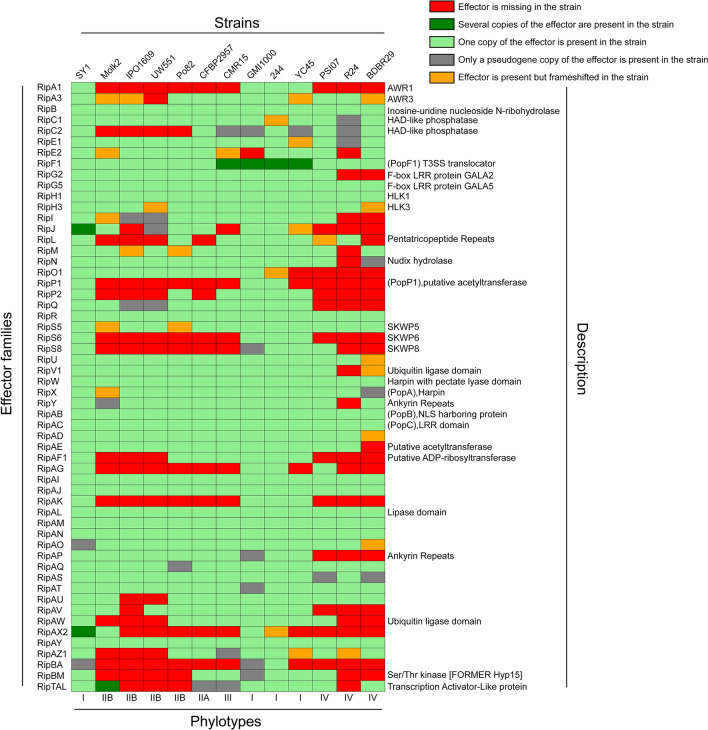
R. solanacearum delivers an array of effector proteins via its type III secretion system (Landry et al. 2020; Xian et al. 2020). Although members of the RSSC can infect many hosts, each strain has its own specific effector proteins, which largely determine their host range (Peeters et al. 2013; Xian et al. 2020). To explore the characteristics of the strain SY1 effector protein arsenal, we analyzed the potential type III effectors of strain SY1 using the T3E database (Peeters et al. 2013). We identified 56 effectors in the strain SY1 genome (Fig. 5), and analyzed the presence of these effector proteins in 12 additional RSSC strains that have been sequenced. The effector proteins of strain SY1 are similar to those of phylotype I strains, such as GMI1000, 224, and YC45, but differ substantially from those of phylotype II and IV strains. In addition, we established that strain SY1 contains more than one copy of the effectors RipJ and RipAX2, but only pseudogene copies of the effectors RipAO and RipBA (Fig. 5). These results demonstrated that SY1 also contents the arsenal of type III effectors, and these effectors has high similarity with that of phylotype I R. solanacearum strains.
Fig. 5.
Identification and comparison of type III effector genes in strain SY1 and the other R. solanacearum strains. The different colored rectangles represent the status of effector protein genes in the corresponding strains. The meanings of the different colors in the rectangles are shown in the legend on the top right. The phylotypes column indicates the phylotype of the different strains. The description column shows the potential physiological or biochemical functions of effector proteins that have been described
Genomics islands and prophage elements
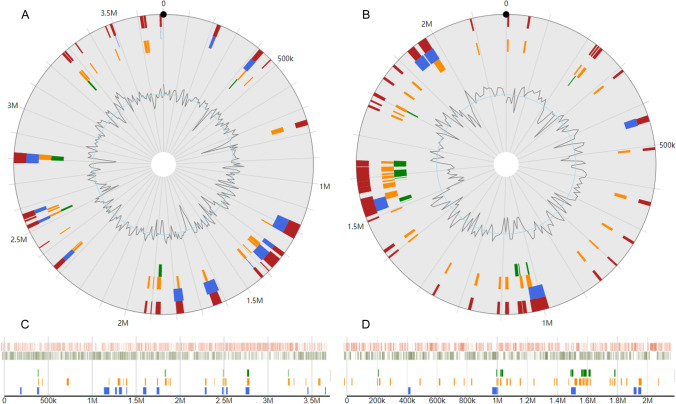
Genomic islands (GIs) are DNA fragments resulting from horizontal gene transfer between different bacterial genomes (Rodriguez-Valera et al. 2016). In this study, we screened the sequences of the chromosome and megaplasmid of strain SY1 for the presence of GIs using Island viewer software (Bertelli et al. 2017). As shown in Fig. 6, 14 and 5 GIs were identified in the chromosome and megaplasmid, respectively, by the IslandPath-DIMOB program. For the chromosome, the lengths of the GIs ranged from 7888 to 60,907 bp, with an average size of 24,200 bp, and a total of 338,806 bp. There were 303 genes in the 14 GIs, which mainly encode transcriptional regulators, transposases, phosphatases, and sugar kinases, for example, l-iditol 2-dehydrogenase, PHB depolymerase family esterase, AAA family ATPase, glutathione S-transferase, FAD-dependent oxidoreductase, and SAM-dependent methyltransferase. For the megaplasmid, the lengths of the GIs ranged from 15,023 to 39,648 bp, with an average size of 25,022 bp, and a total of 125,110 bp. There were 89 genes in the 5 GIs, which mainly encode IS5-like element IS1421 family transposases, LysR family transcriptional regulators, MFS transporters, ABC transporter substrate-binding proteins, transcriptional regulators, mannose-binding lectin proteins, oxidoreductases, AAA family ATPases, and antitoxins.
Fig. 6.
Circular and horizontal plots of genomic islands (GIs) identified in the strain SY1 chromosome (A, circular plot; C, horizontal plot) and megaplasmid (B, circular plot; D, horizontal plot). The orange bars represent the predicted GIs identified by SIGI-HMM, the green bars represent the GIs identified by IslandPick, and the blue bars represent the analysis by IslandPath-DIMOB. The red boxes represent the integrated search results
Accurate identification of prophages is important for the comprehensive study of a pathogen’s genome and its genetic potential (Bondy-Denomy et al. 2016). In this study, using the program Phage_Finder (Fouts 2006), we found that the megaplasmid of the strain SY1 does not have a prophage region that can be predicted, while the chromosome contains two prophage regions (Table S3). These two prophage sequences had a total size of 55,624 bp. The lengths of prophage regions 1 and 2 were 36,895 bp (from 1,188,544 to 1,225,438 bp, with a GC content of 65.87%) and 18,729 bp (from 1,396,400 to 1,415,128 bp, with a GC content of 66.67%), respectively. A total of 78 protein-coding genes were predicted in these prophage sequences of the strain SY1 chromosome, which encoded integrases, RNA-binding proteins, transcriptional regulators, phage capsid scaffolding proteins, and phage tail proteins.
Carbohydrate-active enzymes, antibiotic resistance, and substitution rate (Ka/Ks) analysis
Carbohydrate-active enzymes (CAZymes) are families of enzymes involved in the breakdown of carbohydrates into smaller components by creation, degradation, and modification of glycosidic bonds (Montgomery et al. 2017). In this study, 215 genes in the strain SY1 genome were annotated as CAZyme family genes (Figure S2). Among them, glycosyl transferases and glycoside hydrolases were the most abundant, followed by carbohydrate-binding modules, carbohydrate esterases, auxiliary activities, and polysaccharide lyases.
Furthermore, we analyzed the antibiotic resistance genes (ARGs) in the strain SY1 genome via the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al. 2020) and identified 130 ARGs (Table S4). These ARGs were involved in resistance to fusidic acid, fosfomycin, nitrofurantoin, isoniazid, triclosan, rifampicin, ethambutol, pyrazinamide, streptomycin, and amoxicillin.
To evaluate the evolutionary dynamics, we used the MUMmer 3 package (Hurst 2002) to identify the orthologous genes between these six strains, and the nonsynonymous mutation rate (Ka)/synonymous mutation rate (Ks) ratio of the genes was calculated by the free ratio model (Li et al. 2009). We found four values of Ka/Ks > 0.5 (positive selection), which involved four genes encoding a DUF2244 domain-containing protein, an ATPase, a (2Fe-2S)-binding protein, and a BON domain-containing protein (Table S5).
Discussion
Since the whole genome of R. solanacearum strain GMI1000 was first sequenced in 2002 (Salanoubat et al., 2002), an increasing number of R. solanacearum strains have been sequenced due to advances in sequencing and analysis technologies (Tan et al. 2019; Chen et al. 2021). However, although P. cablin production is seriously threatened by R. solanacearum, there have been few studies on the R. solanacearum strains isolated from P. cablin. In this study, we isolated the strain SY1 from P. cablin and sequenced and analyzed its whole genome, providing an important research basis for studying the characteristics of R. solanacearum in P. cablin, examining the molecular mechanisms of the interaction between R. solanacearum and P. cablin, and developing targeted biological control methods (Wang et al. 2018). One R. solanacearum strain, phylotype I, isolated from a sweet pepper in South China, was been named as Rs-SY1 (Du et al. 2017). Here, the strain named as SY1, although the names look similar, they are two different strains. In this study, we selected five additional virulent R. solanacearum strains, which sequenced and published earlier, and done more research works, GMI1000 (Zhang et al. 2020), EP1 (Li et al. 2016), CMR15 (Ravelomanantsoa et al. 2016), PS107 (Landry et al. 2020), and IPO1609 (González et al. 2011), representing phylotypes I, I, III, IV, and IIB1, respectively. Phylogenetic analysis of strain SY1 and these five strains demonstrated that strain SY1 can be classified into phylotype I, which includes strains mostly from Asia, and this result is consistent with the generally accepted view that the evolutionary relationships among R. solanacearum strains are related to their geographical origin (Peeters et al., 2013). In addition, the Average Nucleotide Identity (ANI) analysis also found that strain SY1 and strain GMI1000 had the highest correlation (Table S6 and S7). However, we established that the strain SY1 has many genes that differ from the two representative strains of phylotype I. KEGG pathway analysis showed that the unique genes of strain SY1 were mainly enriched in the pathways of ‘plant–pathogen interaction’, ‘mismatch repair’, and ‘nucleotide excision repair’, which may function in host-specific infection and survival in certain environments. Noteworthy, four genes were enriched in the pathway of ‘plant–pathogen interaction’ (Table S1). Two of them were annotated as the “HlyD family efflux transporter periplasmic adaptor subunit”, and the other two were annotated as the “ATP-binding cassette domain-containing protein”. These genes were reported plays a key role in the pathogenicity and drug resistance of pathogens (Schmidt and Hensel 2004; Kostakioti et al. 2005). It remains to be determined whether strain SY1 has a stronger ability to infect P. cablin than other R. solanacearum strains, or if it is more suitable for the soil environment of P. cablin production areas.
In this study, we investigated the arsenal of effectors of R. solanacearum strains, which are key to determining the host range of a particular strain (Genin and Denny 2012). Using the T3E database (Peeters et al., 2013), we predicted the potential type III effectors of the strain SY1 and compared them with the effector proteins in 12 other RSSC strains that have been sequenced. Our results were consistent with those of the comparative genomics analysis; the effector proteins of strain SY1 were similar to those of phylotype I strains (which includes strains mostly from Asia) but largely differed from those of strains from other phylotypes. Thus, strain SY1 has an arsenal of effectors that allow for pathogenesis of host plants in Asia. Notably, strain SY1 also has strain-specific type III effectors, such as the effectors encoded by J4H89_06410, J4H89_15630, J4H89_18075, and J4H89_24965 genes (Table S2). Among these, J4H89_06410 and J4H89_24965 encode effectors that are similar to the type III effectors from Pseudomonas amygdali and Burkholderia pseudomallei, respectively (Table S2). We speculated that these may have been obtained through horizontal gene transfer. In addition, although R. solanacearum can infect hundreds of different hosts and has a large pool of effector proteins, specific strains may have a host preference. We noticed that the strain SY1 has more than one copy of the effectors RipJ and RipAX2, but only a pseudogene copy of RipAO and RipBA. The effector proteins of R. solanacearum have functional redundancy, and many effector proteins are also recognized by host immunity (Landry et al. 2020). It is noteworthy that since RipJ belongs to the well-known YopJ family of acetyltransferases but is an avirulence protein that triggers bacterial wilt resistance in Solanum pimpinellifolium (Pandey et al. 2021). These results suggest that the role of these effectors in strain SY1 infection may be amplified or negligible during the infection of some specific host plants.
Conclusions
Here, we sequenced the complete genome of the R. solanacearum strain SY1 isolated from infected P. cablin plants. Comparative genomics analysis of strains of R. solanacearum from different phylotypes demonstrated the genetic diversity and host specificity of these strains. Strain SY1 was classified into phylotype I though it contained 273 unique genes. The analysis of strain-specific genes and type III effectors suggested that these genes and effectors may contribute to the host specificity of the strain SY1. These results extend our knowledge of R. solanacearum genomes, laying the foundation for further studies on the interaction between P. cablin and R. solanacearum and providing a theoretical basis for the prevention and control of bacterial wilt.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Xiaodan Tan and Dr. Yong Yang (Zhongkai University of Agriculture and Engineering, Guangzhou, China) for assisting in the data analysis. We appreciate Guangzhou Genedenovo Biotechnology Co., Ltd. for technical support.
Abbreviations
- RSSC
R. solanacearum Species complex
- T3Es
Type III effector proteins
- NR
Non-redundant protein database
- GIs
Genomics islands
- CAZymes
Carbohydrate-active enzymes
- ARGs
Antibiotic resistance genes
- CARD
Comprehensive antibiotic resistance database
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
Author contributions
YS: conceptualization, data curation, writing—original draft preparation, investigation, supervision, writing—review & editing. YS: conceptualization, supervision, writing—review & editing. AH: conceptualization, supervision, writing—review & editing. LX: methodology, data curation, writing—review & editing. CL: conceptualization, writing—review & editing. JZ: methodology, data curation, writing—review & editing. ZM: software, writing—review & editing. ZD: software, writing—review & editing. GY: supervision, writing—review & editing.
Funding
This work was supported by the National Natural Science Foundation of China (Grant no. 32000086 to Yunhao Sun, for the analysis of the data and writing of the manuscript), the Applied Basic Research Programs of Science and Technology Commission Foundation of Guangdong Province (Grant no. 2019A1515110593 to Yunhao Sun, for the analysis of the data and writing of the manuscript), the Key Scientific and Technological Innovation Projects of Guangdong Forestry Bureau (Grant no. 2020KJCX009 to Yunhao Sun, for the collection of data), the Research Project of Innovative Institute for Plant Health from Zhongkai University of Agriculture and Engineering (Grant no. KA21031H103 to Yunhao Sun, for the collection of data), and the Starting Research Fund from Zhongkai University of Agriculture and Engineering (grant no. KA200540844 to Yunhao Sun, for the collection of data).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. The genome sequencing raw data of Ralstonia solanacearum strain SY1 is available via GenBank under the SRA accessions CP071914 (chromosome) (https://www.ncbi.nlm.nih.gov/nuccore/CP071914.1/) and CP071915 (megaplasmid) (https://www.ncbi.nlm.nih.gov/nuccore/CP071915.1/).
Declarations
Conflict of interests
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhangyong Dong, Email: dongzhangyong@hotmail.com.
Guohui Yu, Email: ygh76411@zhku.edu.cn.
References
- Aisyah Y, Yunita D, Amanda A. Antimicrobial activity of patchouli (Pogostemon cablin Benth) citronella (Cymbopogon nardus), and nutmeg (Myristica fragrans) essential oil and their mixtures against pathogenic and food spoilage microbes. IOP Conf Ser Earth Env Sci. 2021;667:1. doi: 10.1088/1755-1315/667/1/012020. [DOI] [Google Scholar]
- Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–d525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez B, López MM, Biosca EG. Survival strategies and pathogenicity of Ralstonia solanacearum phylotype II subjected to prolonged starvation in environmental water microcosms. Microbiol (read, Engl) 2008;154(Pt 11):3590–3598. doi: 10.1099/mic.0.2008/019448-0. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FSL. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–w35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol. 2014;31(5):1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, Maxwell KL. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016;10(12):2854–2866. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Chen K, Wang L, Chen H, Zhang C, Wang S, Chu P, Li S, Fu H, Sun T, Liu M, et al. Complete genome sequence analysis of the peanut pathogen Ralstonia solanacearum strain Rs-P.362200. BMC Microbiol. 2021;21(1):118. doi: 10.1186/s12866-021-02157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13(12):1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Song ES, Heu S, Baek J, Lee YK, Lee S, Lee SW, Park DS, Lee TH, Kim JG, et al. Prediction of host-specific genes by pan-genome analyses of the Korean Ralstonia solanacearum species complex. Front Microbiol. 2019;10:506. doi: 10.3389/fmicb.2019.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsing BL, Truchon AN, Gonzalez-Orta ET, Milling AS, Allen C. Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen-limited host xylem environment. Mbio. 2015;6(2):e02471. doi: 10.1128/mBio.02471-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du HS, Chen B, Zhang XF, Zhang FL, Miller SA, Rajashekara G, Xu XL, Geng SS. Evaluation of Ralstonia solanacearum infection dynamics in resistant and susceptible pepper lines using bioluminescence imaging. Plant Dis. 2017;101(2):272–278. doi: 10.1094/PDIS-05-16-0714-RE. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34(20):5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin S, Denny TP. Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol. 2012;50:67–89. doi: 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- González A, Plener L, Restrepo S, Boucher C, Genin S. Detection and functional characterization of a large genomic deletion resulting in decreased pathogenicity in Ralstonia solanacearum race 3 biovar 2 strains. Environ Microbiol. 2011;13(12):3172–3185. doi: 10.1111/j.1462-2920.2011.02636.x. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web server issue):W52–57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411(6841):1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genetics. 2002;18(9):486. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Junren C, Xiaofang X, Mengting L, Qiuyun X, Gangmin L, Huiqiong Z, Guanru C, Xin X, Yanpeng Y, Fu P, et al. Pharmacological activities and mechanisms of action of Pogostemon cablin Benth: a review. Chin Med. 2021;16(1):5. doi: 10.1186/s13020-020-00413-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. Mechanisms of protein export across the bacterial outer membrane. J Bacteriol. 2005;187(13):4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry D, González-Fuente M, Deslandes L, Peeters N. The large, diverse, and robust arsenal of Ralstonia solanacearum type III effectors and their in planta functions. Mol Plant Pathol. 2020;21(10):1377–1388. doi: 10.1111/mpp.12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Z, Vang S, Yu J, Wong GK, Wang J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J Mol Evol. 2009;68(4):414–423. doi: 10.1007/s00239-009-9222-9. [DOI] [PubMed] [Google Scholar]
- Li P, Wang D, Yan J, Zhou J, Deng Y, Jiang Z, Cao B, He Z, Zhang L. Genomic analysis of phylotype I strain EP1 reveals substantial divergence from other strains in the Ralstonia solanacearum species complex. Front Microbiol. 2016;7:1719. doi: 10.3389/fmicb.2016.01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QQ, Huo YY, Chen CJ, Zeng ZY, Xu FR, Cheng YX, Dong X. Biological activities of two essential oils from Pogostemon cablin and Eupatorium fortunei and their major components against fungi isolated from Panax notoginseng. Chem Biodivers. 2020;17(12):e2000520. doi: 10.1002/cbdv.202000520. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44(W1):W54–57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6):614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkin I, Pham H, Starostina E, Vyahhi N, Pham S. C-Sibelia: an easy-to-use and highly accurate tool for bacterial genome comparison. F1000Research. 2013;2:258. doi: 10.12688/f1000research.2-258.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery AP, Xiao K, Wang X, Skropeta D, Yu H. Computational glycobiology: mechanistic studies of carbohydrate-active enzymes and implication for inhibitor design. Adv Protein Chem Struct Biol. 2017;109:25–76. doi: 10.1016/bs.apcsb.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinf (oxf, Engl) 2013;29(22):2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Moon H, Choi S, Yoon H, Prokchorchik M, Jayaraman J, Sujeevan R, Kang Y, McCann H, Segonzac C, et al. Ralstonia solanacearum type III effector RipJ triggers bacterial wilt resistance in Solanum pimpinellifolium. Mol Plant Microbe Interact. 2021;34(8):962–972. doi: 10.1094/MPMI-09-20-0256-R. [DOI] [PubMed] [Google Scholar]
- Peeters N, Carrère S, Anisimova M, Plener L, Cazalé AC, Genin S. Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics. 2013;14:859. doi: 10.1186/1471-2164-14-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuwajaroanpong A, Chaniad P, Horata N, Muangchanburee S, Kaewdana K, Punsawad C. In vitro and in vivo antimalarial activities and toxicological assessment of Pogostemon Cablin (Blanco) Benth. J Evid-Based Integr Med. 2020;25:2515690X20978387. doi: 10.1177/2515690X20978387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior P, Ailloud F, Dalsing BL, Remenant B, Sanchez B, Allen C. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics. 2016;17:90. doi: 10.1186/s12864-016-2413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelomanantsoa S, Robène I, Chiroleu F, Guérin F, Poussier S, Pruvost O, Prior P. A novel multilocus variable number tandem repeat analysis typing scheme for African phylotype III strains of the Ralstonia solanacearum species complex. PeerJ. 2016;4:e1949. doi: 10.7717/peerj.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R, Glöckner FO, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2015;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, López-Pérez M. Flexible genomic islands as drivers of genome evolution. Curr Opin Microbiol. 2016;31:154–160. doi: 10.1016/j.mib.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Safni I, Cleenwerck I, De Vos P, Fegan M, Sly L, Kappler U (2014) Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. Nov. Int J Syst Evol Microbiol 64(9):3087–3103 [DOI] [PubMed]
- Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus JC, Cattolico L, et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415(6871):497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Hensel M. Pathogenicity Islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17(1):14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinf (oxf, Engl) 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Silva-Pereira TT, Ikuta CY, Zimpel CK, Camargo NCS, de Souza Filho AF, Ferreira Neto JS, Heinemann MB, Guimarães AMS. Genome sequencing of Mycobacterium pinnipedii strains: genetic characterization and evidence of superinfection in a South American sea lion (Otaria flavescens) BMC Genomics. 2019;20(1):1030. doi: 10.1186/s12864-019-6407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li P, Deng M, Shen D, Dai G, Yao N, Lu Y. The Ralstonia solanacearum effector RipAK suppresses plant hypersensitive response by inhibiting the activity of host catalases. Cell Microbiol. 2017;19:8. doi: 10.1111/cmi.12736. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li P, Shen D, Wei Q, He J, Lu Y. The Ralstonia solanacearum effector RipN suppresses plant PAMP-triggered immunity, localizes to the endoplasmic reticulum and nucleus, and alters the NADH/NAD(+) ratio in Arabidopsis. Mol Plant Pathol. 2019;20(4):533–546. doi: 10.1111/mpp.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Qiu H, Li F, Cheng D, Zheng X, Wang B, Huang M, Li W, Li Y, Sang K, et al. Complete genome sequence of sequevar 14M Ralstonia solanacearum strain HA4-1 reveals novel type III effectors acquired through horizontal gene transfer. Front Microbiol. 2019;10:1893. doi: 10.3389/fmicb.2019.01893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N (2009) Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protocols Bioinf Chapter 4:Unit 4.10 [DOI] [PubMed]
- Tasset C, Bernoux M, Jauneau A, Pouzet C, Brière C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010;6(11):e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij S, Kuhl H, Kuznetsova IS, Komissarov A, Yurchenko AA, Van Heusden P, Singh S, Thevasagayam NM, Prakki SR, Purushothaman K, et al. Chromosomal-level assembly of the Asian seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet. 2016;12(4):e1005954. doi: 10.1371/journal.pgen.1005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinf. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang L, Zhou K, Zhang Y, Song Z, He S. Evidence for adaptation to the Tibetan plateau inferred from Tibetan loach transcriptomes. Genome Biol Evol. 2015;7(11):2970–2982. doi: 10.1093/gbe/evv192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Jin H, Deng Z, Li Z, Mai Y, Li G, He H. A practical random mutagenesis system for Ralstonia solanacearum strains causing bacterial wilt of Pogostemon cablin using Tn5 transposon. World J Microbiol Biotechnol. 2018;35(1):7. doi: 10.1007/s11274-018-2581-x. [DOI] [PubMed] [Google Scholar]
- Xian L, Yu G, Wei Y, Rufian JS, Li Y, Zhuang H, Xue H, Morcillo RJL, Macho AP. A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe. 2020;28(4):548–557. doi: 10.1016/j.chom.2020.07.003. [DOI] [PubMed] [Google Scholar]
- Xiao CL, Chen Y, Xie SQ, Chen KN, Wang Y, Han Y, Luo F, Xie Z. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods. 2017;14(11):1072–1074. doi: 10.1038/nmeth.4432. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li G, Li Q, He L, Zhang Y, Wang Y, He H. Identification and characterization of virulence-attenuated mutants in Ralstonia solanacearum as potential biocontrol agents against bacterial wilt of Pogostemon cablin. Microb Pathog. 2020;147:104418. doi: 10.1016/j.micpath.2020.104418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The genome sequencing raw data of Ralstonia solanacearum strain SY1 is available via GenBank under the SRA accessions CP071914 (chromosome) (https://www.ncbi.nlm.nih.gov/nuccore/CP071914.1/) and CP071915 (megaplasmid) (https://www.ncbi.nlm.nih.gov/nuccore/CP071915.1/).