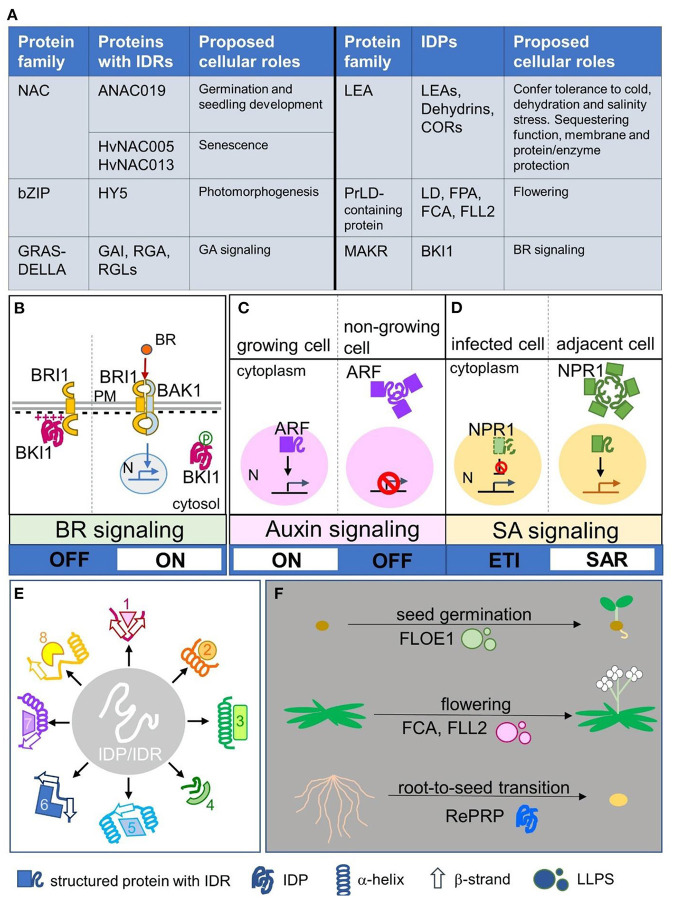
Figure 1.
Plant IDP/IDR members and their spatial regulation, broad specificity, and switch of signaling and physiological status. (A) Typical examples of plant protein families with IDP members and proteins with IDRs, including their proposed cellular roles. (B) A case of spatial regulation of IDPs modulated by electrostatic interaction and post-translational modification. Intrinsically disordered BKI1 associated with the plasma membrane by its disordered cationic region and inhibited the formation of BR receptors (BRI1 and BAK1). Phosphorylation of this region causes electrostatic repulsion to release BKI1 from the plasma membrane into the cytosol; thus BRI1 and BAK1 can form active BR receptors and promote downstream BR signaling. (C,D) Two cases showing spatial regulation of IDRs via forming biomolecular condensates. (C) The nuclear–cytoplasmic compartmentation of ARF mediates auxin signaling turning on or off. In actively growing cells, ARF was present in the nucleus and auxin signaling was activated, whereas in non-growing cells, ARF formed condensates via its IDRs and was retained in the cytoplasm; thus auxin signaling was inactivated. (D) The nuclear–cytoplasmic compartmentation of NPR1 mediates SA signaling to trigger the ETI or SAR program. In infected cells, the ETI program is promoted to activate programmed cell death, whereas the inhibitor NPR1 is degraded in the nucleus. In adjacent cells, nucleus-localized NPR1 triggers the SAR program, whereas cytoplasmic NPR1 forms biomolecular condensates to inhibit proteins involved in programmed cell death. (E) IDPs/IDRs may adopt various secondary conformations such as α-helixes or β-strands when interacting with different partners, which generate broad specificity. (F) Three cases showing that protein phase transition controls physiological phase transition. Disordered FLOE1 senses water content and controls seed germination. Disordered FCA and FLL2 form LLPS to control flowering, a phase transition from the vegetative phase to reproductive phase. Intrinsically disordered rice RePRP enables root-to-seed transition as an adaptation response to water deficit stress. IDPs/IDRs: intrinsically disordered proteins/regions; NAC: NO APICAL MERISTEM, ATAF, CUP-SHAPED COTYLEDON; bZIP: basic domain/leucine zippers; HY5: LONG HYPOCOTYL 5; GRAS: GIBBERELLIC ACID INSENSITIVE, REPRESSOR of GAI, SCARECROW; DELLA: Asp-Glu-Leu–Leu-Ala; GAI: GIBBERELLIC ACID INSENSITIVE; RGA: REPRESSOR of GAI; RGL: RGA-LIKE; GA: Gibberellic acid; LEA: LATE EMBRYOGENESIS ABUNDANT; COR: COLD-REGULATED; PrLD: Prion-like domain; LD: Luminidependens; FPA: FLOWERING LOCUS PA; FCA: FLOWERING LOCUS CA; FLL2: FLX-LIKE 2; MAKR: MEMBRANE-ASSOCIATED KINASE REGULATOR; BKI1: BRI1 KINASE INHIBITOR1; BR: brassinosteroid; BRI1: BRASSINOSTEROID INSENSITIVE 1; BAK1: BRI1-ASSOCIATED RECEPTOR KINASE 1; N: nucleus; PM: plasma membrane; ARF: AUXIN RESPONSE FACTOR; NPR1: NONEXPRESSOR OF PATHOGENESIS-RELATED GENE 1; SA: salicylic acid; SAR: systemic acquired resistance; ETI: effector triggered immunity; LLPS: liquid–liquid phase separation; RePRP: REPETITIVE PROLINE-RICE PROTEIN; 1-8 in (E) represent different putative interacting partners.