Figure 2. NTRAS operates as splicing‐regulatory lncRNA.
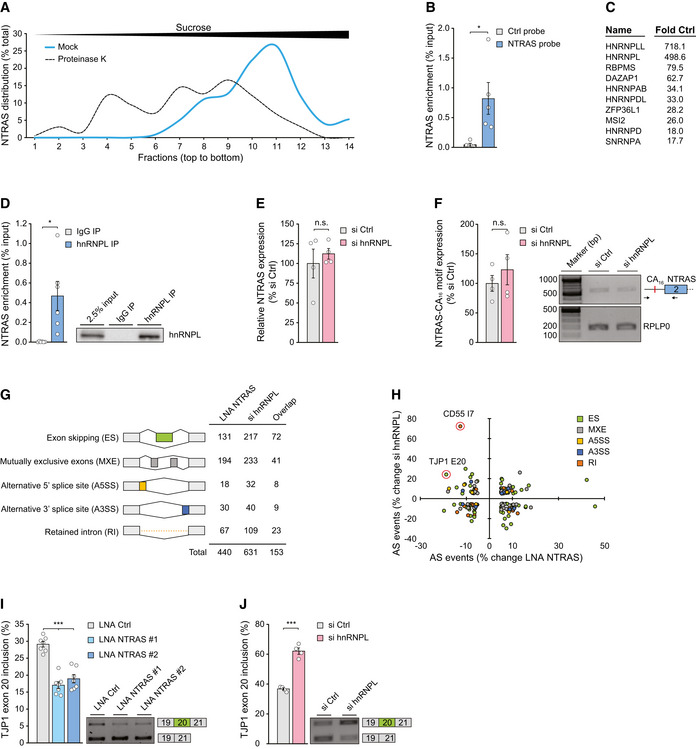
- Separation of mock and proteinase K‐treated HeLa nuclear extracts by sucrose density gradient ultracentrifugation followed by NTRAS detection using RT–qPCR. Fractions 1 and 14 represent top and bottom of the gradient, respectively (n = 1).
- Enrichment of NTRAS by antisense affinity selection from HeLa cell lysate followed by RT–qPCR, comparing a control and an NTRAS‐specific probe (n = 5 independent biological replicates).
- Top ten enriched proteins from NTRAS affinity selections, identified by mass spectrometry (n = 5 independent biological replicates).
- RT–qPCR‐based validation of NTRAS–hnRNPL interaction by anti‐hnRNPL RIPs from HUVEC nuclear fractions (n = 6 independent biological replicates). Representative western blot on the right.
- RT–qPCR analysis of NTRAS expression in HUVECs upon silencing of hnRNPL (n = 4 independent biological replicates).
- RPLP0‐normalized NTRAS‐CA16 motif expression after hnRNPL silencing determined by RT‐PCR (n = 4 independent biological replicates).
- RNA deep sequencing‐based assignment of alternative splicing events in NTRAS‐ or hnRNPL‐silenced HUVECs, using rMATS software. Displayed are changes > 5% (n = 2 independent biological replicates).
- NTRAS and hnRNPL co‐regulated alternative splicing (AS) events. Data points with an FDR < 0.05 are circled (n = 2 independent biological replicates).
- RT–PCR‐based analysis of TJP1 exon 20 inclusion upon silencing of NTRAS in HUVECs (n = 7 independent biological replicates). Representative agarose gels on the right.
- RT–PCR‐based analysis of TJP1 exon 20 inclusion upon silencing of hnRNPL in HUVECs (n = 4 independent biological replicates). Representative agarose gels on the right.
Data information: In (B, D–F, I, J), data are represented as mean ± SEM. n.s.: non‐significant, *P < 0.05, ***P < 0.001. (B–F, I, J) two‐tailed unpaired t‐test.
Source data are available online for this figure.
