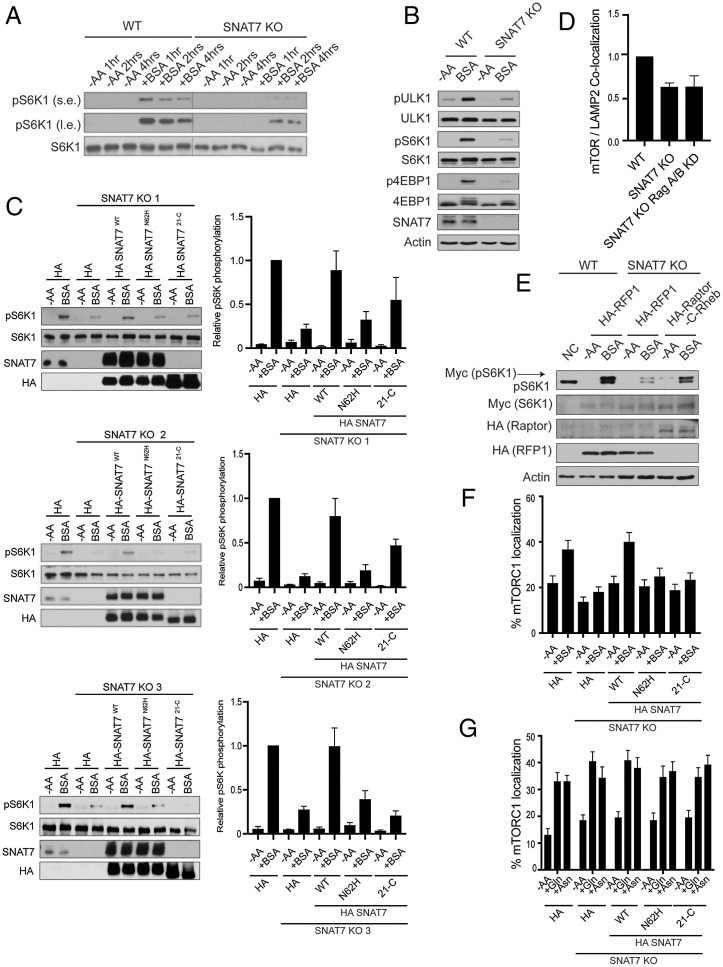
Fig. 3.
SNAT7 is required for albumin-induced mTORC1 activation. (A) SNAT7 deletion impairs mTORC1 activation by macropinocytosis at multiple time points. WT or SNAT7 KO MIA PaCa-2 cells (SNAT7 KO) were starved of amino acids for 2 h and then maintained in amino acid starvation media (−AA) or stimulated with 3% bovine serum albumin (+BSA) for the indicated times. mTORC1 activity was analyzed by immunoblotting for the phosphorylation status of S6K1 (pS6K1) at threonine 389. s.e., short exposure; l.e., long exposure. S6K1 was probed for as a loading control. (B) SNAT7 is required for macropinocytosis mediated mTORC1 activation of multiple downstream substrates. SNAT7 KO MIA PaCa-2 cells were starved of serum and amino acids (−AA) and stimulated with 3% BSA for 4 h. mTORC1 activity was analyzed by immunoblotting for the phosphorylation status of ULK1 at serine 758 (pULK1), S6K1 at threonine 389 (pS6K1), 4EBP1 at serine 65 (p4EBP1), or by mobility shift of the total 4EBP1 protein. ULK1, S6K1, SNAT7, and actin were probed for as controls. (C) SNAT7 is required for BSA-induced mTORC1 activation. WT MIA PaCa-2 cells, SNAT7 KO (3 different clones as indicated), or SNAT7 KO cells overexpressing HA-tagged SNAT7WT, HA-tagged SNAT7N62H, or HA-tagged SNAT721-C were starved of amino acids (−AA) for 2 h and then stimulated with BSA. (Left) mTORC1 activity was analyzed by the phosphorylation of S6K. SNAT7 levels were confirmed using Western blotting. S6K and HA are loading controls. (Right) Quantification of pS6K levels and statistical analysis. Clone 1: WT + BSA vs. SNAT7 KO + BSA, P < 0.001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.001; WT + BSA vs. HA-SNAT721-C + BSA, P > 0.05; and SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.05. Clone 2: WT + BSA vs. SNAT7 KO + BSA, P < 0.0001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.001; WT + BSA vs. HA-SNAT721-C + BSA, P < 0.01; and SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.05. Clone 3: WT + BSA vs. SNAT7 KO + BSA, P < 0.0001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.01; WT + BSA vs. HA-SNAT721-C + BSA, P < 0.001; and SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.05. (D) SNAT7 regulates mTORC1 lysosomal localization. WT, SNAT7 KO, or SNAT7 KO with RagA/B knocked down (KD) MIA PaCa-2 cells were assessed for mTORC1 lysosomal localization. Images were quantified for mTOR/LAMP2 colocalization. Significance (P value): WT vs. SNAT7 KO, P < 0.01; WT vs. SNAT7 KO Rag A/B KD, P < 0.05; and SNAT7 KO vs. SNAT7 KO Rag A/B KD, P = 0.984. (E) mTORC1 activation deficiency in SNAT7 KO cells can be rescued by artificially targeting mTORC1 to the lysosome. WT or KO MIA PaCa-2 cells were cotransfected with HA-tagged RFP1, Raptor, or Raptor fused with the C terminus of Rheb (Raptor-C-Rheb), together with Myc-tagged S6K1. Cells were starved of amino acids (−AA) for 2 h and stimulated with 3% BSA for 4 h. mTORC1 activity was assessed as in (A). Myc, HA, and actin were probed for as controls. (F) SNAT7 is required for BSA-induced mTORC1 lysosomal localization. WT, SNAT7 KO, or SNAT7 KO cells overexpressing HA-tagged SNAT7WT, HA-tagged SNAT7N62H, or HA-tagged SNAT721-C were starved of amino acids (−AA) for 2 h and then stimulated with 3% BSA for 4 h. Colocalization of mTOR and the lysosomal protein (LAMP2) was analyzed via confocal microscopy. P values: WT -AA vs. WT + BSA, P < 0.05; HA-SNAT7WT −AA vs. HA-SNAT7WT + BSA, P < 0.01; WT + BSA vs. SNAT7 KO + BSA, P < 0.001; SNAT7 KO + BSA vs. HA-SNAT7WT + BSA, P < 0.001; WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.05; WT + BSA vs. HA-SNAT721-C + BSA, P < 0.05; SNAT7WT + BSA vs. HA-SNAT7N62H + BSA, P < 0.05; and SNAT7WT + BSA vs. HA-SNAT721-C + BSA, P < 0.01. (G) SNAT7 is not required for glutamine and asparagine induced mTORC1 lysosomal localization. WT, SNAT7 knockout (KO), or SNAT7 KO MIA PaCa-2 cells overexpressing HA-tagged SNAT7WT, HA-tagged SNAT7N62H, or HA-tagged SNAT721-C were starved of amino acids (−AA) for 2 h and then stimulated with 4 mM glutamine (+Gln) or asparagine (+Asn) for 2 h. mTOR and lysosomal protein (LAMP2) colocalization was analyzed with confocal microscopy. P values: WT −AA vs. WT + Gln, P < 0.001; WT −AA vs. WT + Asn, P < 0.001; SNAT7 KO −AA vs. SNAT7 KO + Gln, P < 0.001; SNAT7 KO −AA vs. SNAT7 KO +Asn, P < 0.01; SNAT7WT −AA vs. SNAT7WT + Gln, P < 0.001; SNAT7WT −AA vs. SNAT7WT + Asn, P < 0.001; HA-SNAT7N62H −AA vs. HA-SNAT7N62H + Gln, P < 0.01; HA-SNAT7N62H −AA vs. HA-SNAT7N62H + Asn, P < 0.01; HA-SNAT721-C −AA vs. HA-SNAT721-C + Gln, P < 0.01; and HA-SNAT721-C −AA vs. HA-SNAT721-C + Asn, P < 0.001.