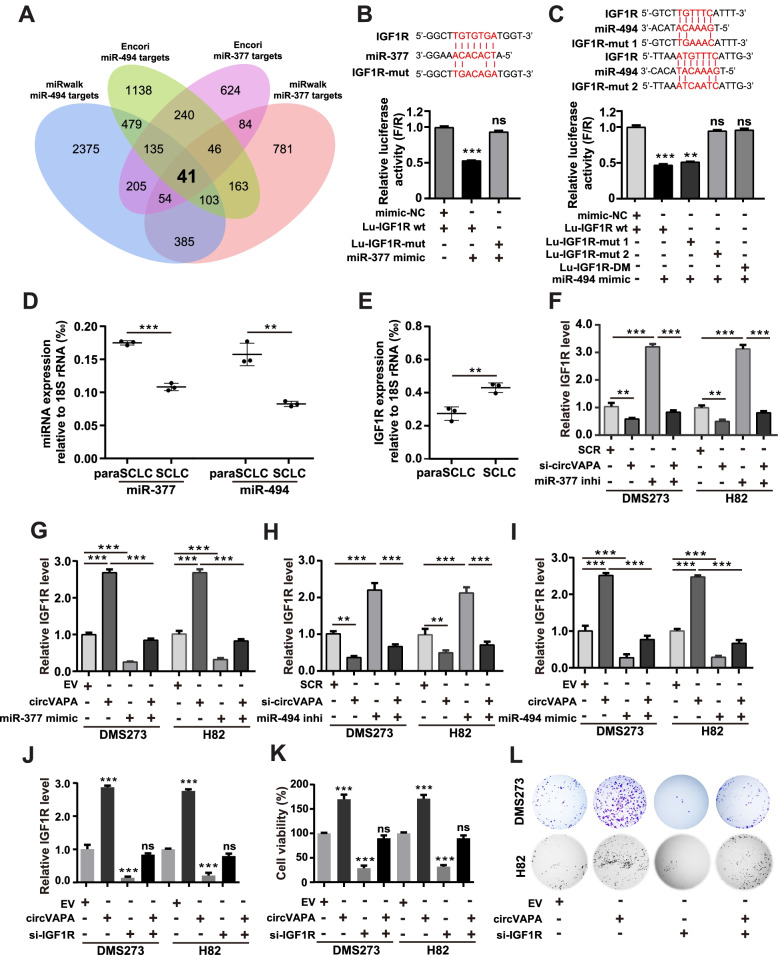
Fig. 5.
circVAPA facilitates SCLC cell viability via the miR-377-3p & miR-494-3p/IGF1R/AKT axis. a Two databases (miRwalk and ENCORI) were used to predict the potential target mRNAs of miR-377-3p and miR-494-3p. The Venn diagram shows the number of overlapping miRNAs. b and c Dual-luciferase reporter assay was performed to validate the interaction of IGF1R and miR-377-3p (b) or miR-494-3p (c). 293T cells were co-transfected with a dual-luciferase reporter vector containing the putative binding sites/ mutated sequences for IGF1R and miR-377-3p/miR-494-3p, miR-377-3p/miR-494-3p mimics or negative control mimics (mimic-NC), respectively. Lu-IGF1R-DM represents Lu-IGF1R-Mut 1 & 2, which contains two mutation sites. d-e RT-qPCR analysis of miR-377-3p/miR-494-3p (d) and IGF1R (e) in 3-paired SCLC and paraSCLC samples. 18S rRNA was used as the internal control. f-j RT-qPCR analysis of IGF1R expression in SCLC cells transiently transfected with siRNAs or plasmids as indicated. k and l Cell viability (k) and colony formation (l) assays of SCLC cells transiently transfected with siRNAs, plasmids, miRNA inhibitors or mimics as indicated. SCR, siRNA with scrambled sequences; si-circVAPA, the co-transfection of two independent siRNAs target circVAPA; EV, the empty vector; circVAPA, the circVAPA overexpression plasmid; miR-377 mimic/miR-494 mimic, transiently overexpressing miR-377-3p/miR-494-3p, respectively; miR-377 inhibitor/miR-494 inhibitor, transiently suppressing miR-377-3p/miR-494-3p, respectively; si-IGF1R, an independent siRNA targeting IGF1R. (All data are presented as the mean ± SD; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed Student’s t-test). Three independent assays were performed in the above assays. *** miR-377/miR-494 represents miR-377-3p/miR-494-3p, respectively