Extended Data Fig. 10. Validation of genetic and molecular reagents for Npnt mutant analysis.
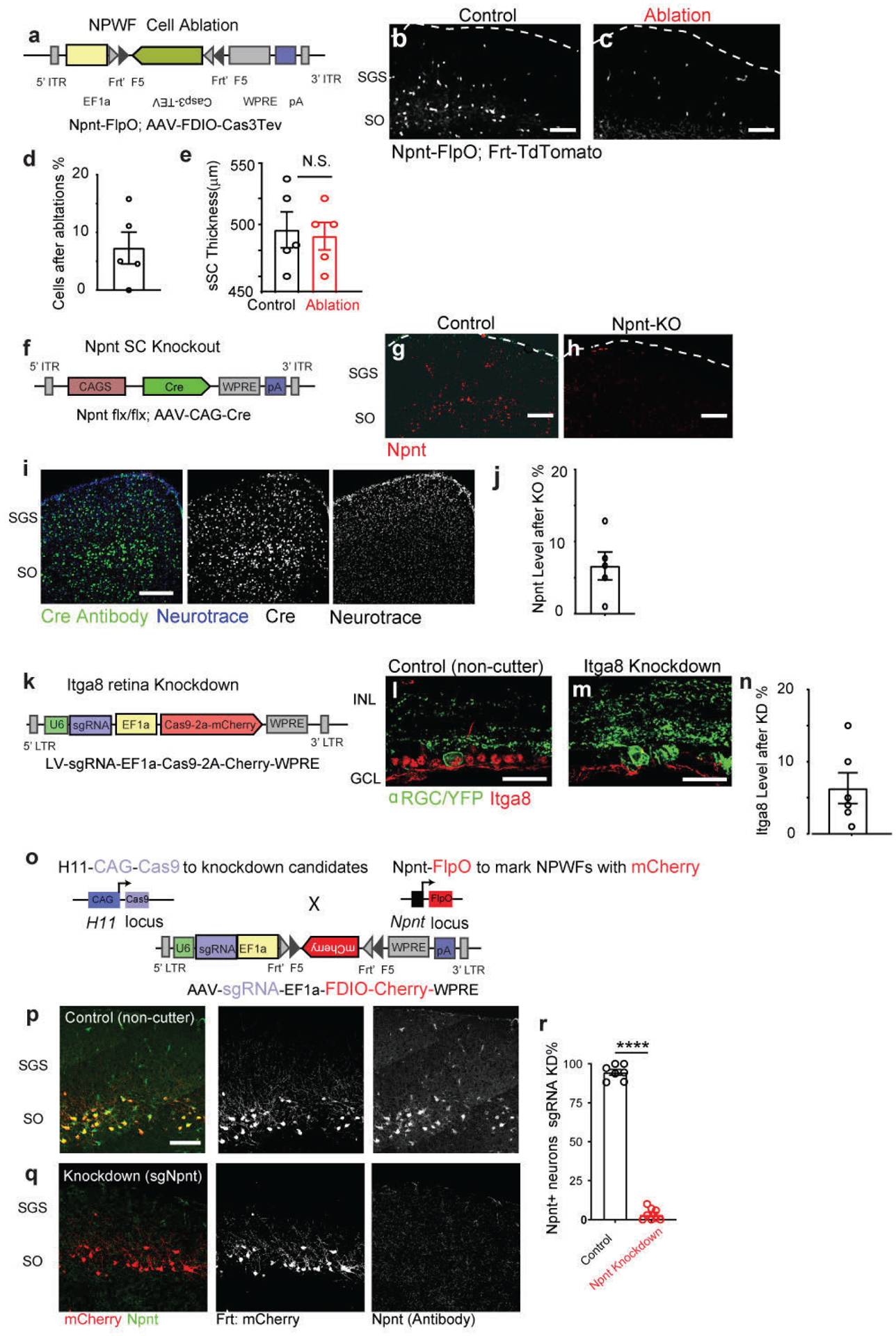
a, The AAV vector design for FlpO-dependent Caspase3-TEV expression, modified from (Addgene #45580)30. b, c, Confirmation of successfully eliminating ESC1s using AAV-fDIO-Caspase3 in Npnt-FlpO by comparing control b and Cas3-TEV injections c. The dotted line marks the pial surface. Scale bar: 100μm. d, Quantifications of Cas3-deletion of NPWFs efficiencies, compared to control side. N=5 animals. e, Quantifications of the superficial SC thickness subject to NPWF eliminations. n=5 animals, per genotype, N.S., no significance, two-sided Student’s t-test. f, The AAV vector design for Cre overexpression. g-h, Confirmation of SC-specific knockout of Npnt using Npnt f/f conditional mutants by comparing controls (g) and mutants (h), n=3 animals. Scale bar: 100μm. No obvious cell-autonomous migration or morphological changes of labeled wide-field neurons were observed after Npnt knockout. i. Sample images showing comprehensive Cre coverage across different sublaminae of SC subject to neonatal AAV injections. Anti-Cre staining was displayed here in the green channel. j, Quantifications of AAV-Cre mediated Npnt knockout efficiency, compared to the control side. n=5 animals. k, The lentiviral vector design for Itga8 knockout based on CRISPR-V2-system to simultaneously express sgRNA/Cas9-2a-Cherry (Addgene Plasmid #99154). l, m, Confirmation of the lentivirus-mediated Itga8 knockout in the retina (m) compared with control (l). Scale bar: 50μm. n, Quantifications of sgRNA-mediated retinal Itga8 knockdown efficacies, compared to control side. n=6 animals, Several sgRNAs were evaluated, and an efficient sgRNA targeting the fourth exon of Npnt was selected (See Methods). Cas9/sgRNA-mediated Npnt deletion was validated by immunostaining with Npnt antibody. We performed neonatal injections of AAV-encoding for sgRNA targeting Npnt or control sgRNA-non-cutter and Flp-dependent RFP expression (AAV-sgNpnt-FDIO-mCherry; or AAV-sg-Noncutter-FDIO-mCherry;) into Kcng4-YFP; Npnt-FlpO; H11Cas9 mice. o, The AAV vector is carrying sgRNAs and Frt-dependent mCherry. This vector can simultaneously knock down endogenous Npnt while labeling the NPWF neurons using Npnt-FlpO dependent RFP reporter. The Cas9 was genetically harbored in the H11-Cas9 line via a genetic cross to Npnt-FlpO. p, q, Npnt in the SC was knocked down (q) compared to sg-Non-cutter control (p). Scale bar: 100μm. The percentages of Npnt knockdown were quantified in (r), n=4 animals, per genotype, ****. p<0.0001, two-sided Student’s T-test. All data in this figure are presented as mean ± SEM.
