Significance
Understanding how protein scaffolds direct mineral morphogenesis is crucial for engineering bone and tooth and would open new vistas in materials design. In the case of tooth enamel, which is the hardest tissue in the body and consists of organized bundles of coaligned apatite crystals, amyloid-like amelogenin nanoribbons are hypothesized to provide the scaffold. We show that these nanoribbons are far more potent calcium phosphate nucleators than other amelogenin motifs or collagen, which provides the scaffold for bone. This potency stems from a periodic array of charged sites that provide a template for calcium phosphate ion binding on a low-energy interface. The ubiquity of β-sheet protein structures suggests that this mechanism can be adopted for the design of synthetic mineralization-directing scaffolds.
Keywords: β-sheet, amelogenin, nucleation, scaffold, calcium phosphate
Abstract
Protein scaffolds direct the organization of amorphous precursors that transform into mineralized tissues, but the templating mechanism remains elusive. Motivated by models for the biomineralization of tooth enamel, wherein amyloid-like amelogenin nanoribbons guide the mineralization of apatite filaments, we investigated the impact of nanoribbon structure, sequence, and chemistry on amorphous calcium phosphate (ACP) nucleation. Using full-length human amelogenin and peptide analogs with an amyloid-like domain, films of β-sheet nanoribbons were self-assembled on graphite and characterized by in situ atomic force microscopy and molecular dynamics simulations. All sequences substantially reduce nucleation barriers for ACP by creating low-energy interfaces, while phosphoserines along the length of the nanoribbons dramatically enhance kinetic factors associated with ion binding. Furthermore, the distribution of negatively charged residues along the nanoribbons presents a potential match to the Ca–Ca distances of the multi-ion complexes that constitute ACP. These findings show that amyloid-like amelogenin nanoribbons provide potent scaffolds for ACP mineralization by presenting energetically and stereochemically favorable templates of calcium phosphate ion binding and suggest enhanced surface wetting toward calcium phosphates in general.
Hierarchical organic–inorganic hybrid structures, such as in tooth enamel and bone, are ubiquitous in biominerals yet challenging to synthesize in vitro. In tooth enamel, mineralization is thought to begin with the nucleation of amorphous calcium phosphate (ACP) in an amelogenin (Amel)-rich extracellular protein matrix, with the ACP transforming into ribbon-like crystals that evolve to highly aligned bundles of apatite (AP) fibers with less than 2% residual protein in the mature stage (1–3). Numerous studies on mineralization by Amel, an intrinsically disordered protein, start from a widely explored model in which Amel assembles into supramolecular nanospheres. However, the pathway by which these nanospheres assemble into a scaffold of bundled fibers that template discrete AP filaments is unclear (4–8). In contrast, recent reports demonstrate that full-length Amel can adopt an amyloid-like (cross-β-sheet) quaternary structure and self-assemble into nanoribbons (NRs) 17 nm wide and 3 to 4 nm in thickness (9, 10), thus providing a conceptually simple scaffold for AP filament formation. Supporting this model, in vivo studies revealed the presence of highly aligned bundles of ribbon-like β-sheet protein assemblies in the developing enamel matrix of various mammals, including humans (9, 11–17), and these NRs match the morphology and quaternary structure of ribbons assembled in vitro from both recombinant human Amel (rH174) (18–20) and synthetic peptides (9). In vivo observations suggest that Amel NRs are involved in the mineralization of AP filaments that form during the secretory stage of amelogenesis (14), while recent in vitro experiments demonstrate that synthetic NRs can indeed template the growth of AP filaments starting with an amorphous precursor formed in the presence of acidic macromolecules (13). However, the extent to which Amel NRs facilitate ACP nucleation, the mechanism that leads to NR templating of ACP, and the basis of that mechanism in the relationship between NR and mineral structure remain unknown.
To address these knowledge gaps, we performed in situ atomic force microscopy (AFM) analyses of calcium phosphate nucleation on self-assembled NRs of the full-length protein, as well as evolutionarily conserved subsegments hypothesized to be important for 1) self-assembly, 2) stabilizing ACP, i.e., phosphoserine-16, and 3) binding to AP, i.e., the C terminus, which is cleaved by matrix metalloproteinase-20 (MMP20) during amelogenesis at the site indicated in Fig. 1A (9, 21–23). To further understand the underlying chemical interactions and correlate NR and mineral structures with energetics, we performed molecular dynamics (MD) simulations of the conformational states of NRs formed from these peptide subsegments using the INTERFACE force field (IFF)/Chemistry at Harvard Macromolecular Mechanics force field (CHARMM36) and compared them with the growth unit of ACP (the Ca2(HPO4)3)2−dimer).
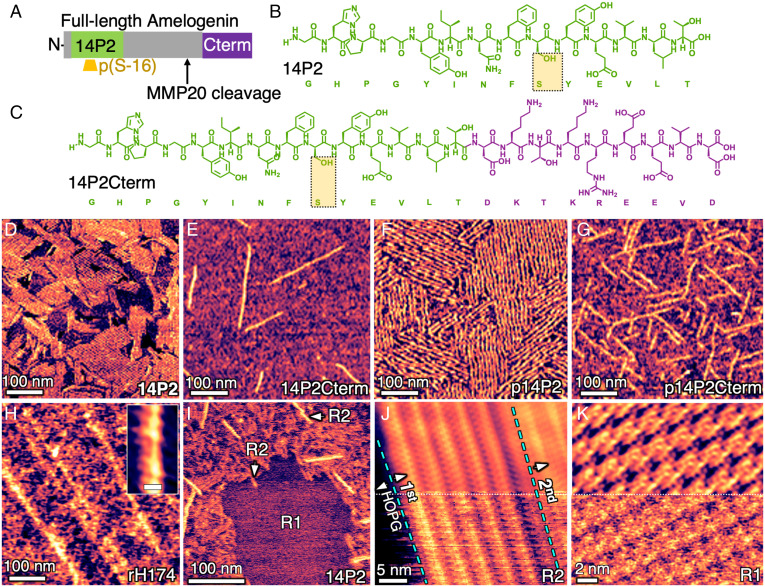
Fig. 1.
Sequences and assemblies of full-length Amel and peptide analogs on HOPG. (A) Location of functional domains in human Amel, (B) self-assembling 14P2 domain, (C) and 14P2Cterm (14P2 appended with truncated C terminus, which is important for AP binding). (D–H) In situ AFM images of ordered NRs assembled on HOPG using 0.1 mg/mL diluted from 1 mg/mL aged for 48 h and characterized in pure water: (D) 14P2; (E) 14P2Cterm; (F) and (G) phosphorylated versions p14P2 and p14P2Cterm, respectively; (H) recombinant full-length Amel, rH174; inset shows higher magnification of ribbon structure reconstructed using 2-dimensional (2D) fast-Fourier transform (FFT) filter (Scale bar, 10 nm). (I) In situ AFM on HOPG in fresh 0.01 mg/mL solution of 14P2 at pH 1.94 reveals 100% coverage with multiple layers: two types, R1 and R2. (J) Structure of R2 in fresh 0.05 mg/mL solution of 14P2 solution at pH 1.94; dashed blue lines delineate the boundary of a second layer over the first on HOPG. Bottom: original image; Top: after reconstruction with 2D FFT filter. (K) Structure of R1 in indicated region of (I). Bottom: original image; Top: after reconstruction with 2D FFT filter.
Based on the above hypotheses, five sequences were designed. Three were nonphosphorylated sequences: one recombinant full-length human Amel (rH174) and two peptide analogs consisting of the domain that drives self-assembly (14P2) and this 14P2 domain with the truncated C terminus appended to it (14P2Cterm), as shown in Fig. 1 A–C. Phosphorylated versions of both peptides, p14P2 and p14P2Cterm, were also synthesized with phosphoserine sites as highlighted in Fig. 1 A–C. The variable, histidine- and proline-rich central domain and other segments of Amel may also play a role in self-assembly or mineralization (23, 24); however, their structures and functions are unclear and were not investigated here.
Results and Discussion
Large Films of NRs.
The previously reported in vitro protocol for NR self-assembly is unsuitable for in situ AFM analyses of calcium phosphate nucleation because it leads to multiple NR polymorphs and has preexisting calcium and phosphate salts in solution that preclude measurements of nucleation rates at known supersaturations. Therefore, the protocol was modified to exclude these salts and polymorphs while promoting the assembly of Amel NRs. In this study, NRs were generated from 0.1 mg/mL solutions taken from 1 mg/mL stock solutions aged for 48 to 336 h at pH 1.94 (see Materials and Methods). With this protocol, all sequences were observed to assemble into NRs both in solution and on the surface of highly ordered pyrolytic graphite (HOPG) and remain stable in water (Fig. 1 D–H) over a pH range of 1.94 to 7.4. While the NR dimensions for each sequence (Table 1) were distinct, the self-assembled NR films were similar in morphology, consisting of individual NRs or NR arrays with well-aligned rows, although the film morphology was unclear in some areas.
Table 1.
Dimensions of NRs measured from in situ AFM images shown in Fig. 1 and SI Appendix, Fig. S2
| Sequence | NR type | Height (nm) | Width (nm) |
|---|---|---|---|
| 14P2 | R2 | 0.65 ± 0.04 | 5.81 ± 0.60 |
| R1 | 0.33 ± 0.07 | 2.51 ± 0.47 | |
| p14P2 | R2 | 1.11 ± 0.1 | 7.57 ± 0.5 |
| R1 | 0.75 ± 0.08 | 3.64 ± 1 | |
| 14P2Cterm | R2 | 0.67 ± 0.08 | 6.16 ± 1.51 |
| R1 | 0.30 ± 0.04 | 3.53 ± 0.36 | |
| p14P2Cterm | R2 | 1.36 ± 0.26 | 9.35 ± 1.24 |
| R1 | 0.82 ± 0.05 | 4.08 ± 0.36 | |
| rH174 | R2 | 0.63 ± 0.05 | 15.00 ± 2.10 |
Error is SD, n ≥ 5.
Higher-magnification analysis of the baseline sequence 14P2 showed that, at a sufficiently high concentration (fresh 0.01 mg/mL solution), HOPG was indeed completely covered, supporting NR layers of two types, referred to as R1 and R2 (Fig. 1I), both of which were in direct contact with HOPG. Of the two, R2 was by far the dominant morphology and formed multiple layers at a sufficiently high concentration (≥ 0.05 mg/mL) without any involvement of R1 NRs (Fig. 1J). In contrast, R1, which was rarely observed for the peptides and was never observed for rH174, only formed single highly crystalline layers and was only observed at concentrations ≤ 0.01 mg/mL (Fig. 1K). In all cases, the NRs were aligned with the three HOPG directions (Fig. 1F and SI Appendix, Fig. S1).
Because all mineralization experiments were performed with R2 NRs, further discussion of NR structure is restricted to this type, although basic morphological information on the R1 layers can be found in Table 1 and the SI Appendix, Fig. S2. The width and height of peptide R2 NRs ranged from 5.81 ± 0.60 nm to 9.35 ± 1.24 nm and from 0.65 ± 0.04 nm to 1.36 ± 0.26 nm, respectively, depending on the sequence (Table 1). For rH174, the measured R2 NR width and height were 15 ± 2.1 nm and 0.63 ± 0.05 nm, respectively. (Note that a 0.5 to 1.0 nm spread in dimensions measured on soft matter, particularly for features below 2 nm, is common due to variations in tip-sample interactions; see SI Appendix, Method 1 for details.)
The dimensions of the R2 NRs are consistent with those of the β-sheet precursors of cross-β-sheet NRs. At 0.5 to 2.0 nm, their thickness is roughly half that of cross-β-sheets depending on the sequence investigated and the AFM substrate and imaging parameters used (25–27). Because β-sheet NRs typically have a hydrophilic and hydrophobic interface on opposing faces, they self-assemble in an anti-parallel fashion on the two hydrophobic interfaces to form the cross-β-sheet structure, which is the motif of in vivo Amel NRs. Here, the HOPG surface is expected to provide one of the hydrophobic surfaces, leading to the formation of β-sheet NRs rather than cross-β-sheets but maintaining the same NR–solution interface as obtained with cross-β-sheet ribbons.
The similarity between the structure and homology of NRs assembled at pH 1.94 and those of previously reported Amel NRs assembled at various pH values, as well as their integrity upon binding to HOPG, was further assessed by a combination of AFM and MD simulations, supported by in situ synchrotron X-ray diffraction (XRD). Deconvolution of the high-resolution structure of the R2-type NRs of 14P2 on HOPG (Fig. 2A) revealed distinct 0.4 to 0.6 nm periodicities at the center of the NRs (Fig. 2B, Middle), consistent with the ∼0.47 nm d-spacing of the β-sheet backbone obtained from bulk solution XRD measurements with or without graphite (SI Appendix, Figs. S3 and S4) and previous reports on 14P2 and rH174 NRs (19, 20, 28). For detailed methods of deconvolution and XRD, see SI Appendix, Methods 2 and 3, respectively. (Note that in contrast to the R2 NR, the measured periodicities of the R1 NR do not match any β-sheet models or XRD d-spacings.)
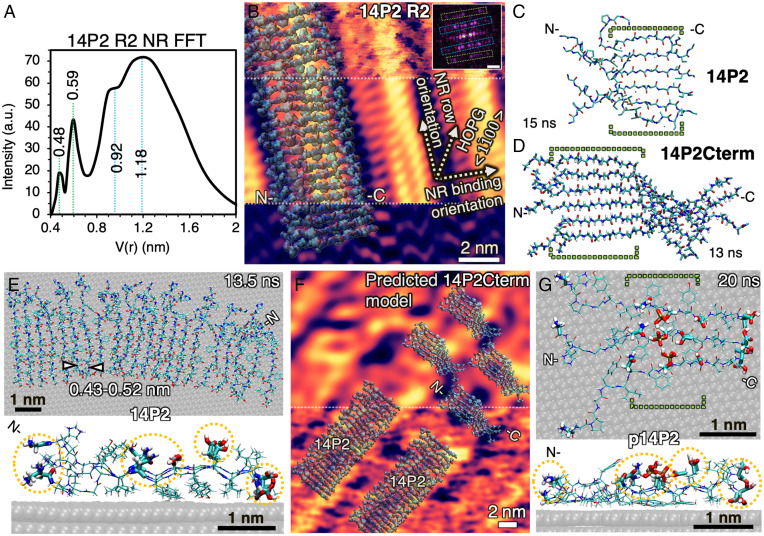
Fig. 2.
Lowest-energy β-sheet conformations of peptides in solution and on HOPG (0001) facet at pH 1.94 from all-atom MD simulations. (A) Radially integrated FFT of 14P2 R2 NR structure on HOPG (Inset in B) by AFM shows distinct ∼0.5 and ∼1 nm periodicities. a.u. is arbitrary unit. (B) 21-monomer β-sheet of 14P2 overlaid on high-resolution AFM structure of 14P2 R2 NRs. Inset FFT shows high-intensity points (in magenta) used to deconvolute the raw image at Top; Middle, 2D-FFT filtered β-sheet structure with ∼0.5 nm periodicities (green boxes in FFT) and without noise and 0.92 to 1.18 nm features; Bottom, only periodic 0.92 to 1.18 nm features (blue boxes in FFT), with highest intensity at edges of the NR (FFT scale bar, 1 nm−1). (C) Snapshot of lowest-energy backbone conformation for 6-monomer β-sheet of 14P2 in bulk solution simulated for 15 ns. (D) Snapshot of 14P2Cterm β-sheet conformation in bulk solution simulated for 13 ns. See SI Appendix, Method 4.3 for details on energies. (E) Top view and side view of 21-monomer 14P2 β-sheet on HOPG simulated for 13.5 ns shows that structure is stable. (F) Comparison of 14P2 model in (B) and simulated 14P2Cterm in (D) overlaid on AFM image of 14P2Cterm; Bottom: original image; Top: 2D-FFT filtered. (G) Top view and side view of a 6-monomer simulation (only 3 displayed) for p14P2 β-sheet on HOPG after 20 ns. Water hidden for clarity in all images. All sequences on graphite have a well-ordered central domain (YINFSY, green brackets in C, D, and G) and hydrophilic sites (thicker bond lines in E and G highlighted by yellow circles), including phosphoserine, that face the solution. Colors of atoms in B to G: cyan is carbon, red is oxygen, blue is nitrogen, white is hydrogen, brown is phosphorous. N- is N terminus and C- is C terminus.
Conformation and Interfacial Chemistry of NRs Used for Mineralization.
To gain insights into the identity and distribution of exposed residues arising from the R2 NR assembly, we performed MD simulations using the INTERFACE Force Field (IFF) in combination with the Chemistry at Harvard Macromolecular Mechanics force field (CHARMM36) with more than 50 conformations for all peptides in solution and on HOPG, including the use of virtual π electrons on graphite and in aromatic amino acids, which was shown to be critical to reproducing solvent and organic interfacial interactions (29–31). Systematic simulations for 13 to 15 ns predicted the stable β-sheet conformations in solution at pH 1.94 for both the basic 14P2 sequence (Fig. 2C) and the three modified versions (Fig. 2D and SI Appendix, Fig. S5). The equilibrated structures of the monomers, which exhibit parallel orientation, showed that YINFSY domains for all sequences were periodic (indicated by dashed green brackets in Fig. 2 C and D), while the N terminus, which was relatively flexible, displayed transient pairing of adjacent N to N termini in a single β-sheet. In contrast, the Cterm domains (DKTKREEVD) in the 14P2Cterm and p14P2Cterm were flexible, tended to twist away from the backbone, and showed pairing of the adjacent C to C termini in a single β-sheet.
On graphite, the calculated energies of adsorption for a single monomer of all sequences range from −23 to −26 ± 8 kcal/mol, indicating that binding is energetically favorable (SI Appendix, Method 4.4). The experimentally observed shortest length of the 14P2 R2 NR was ∼10 nm. Therefore, an equivalent length of a 14P2 β-sheet with 21 monomers was placed on graphite based on the R2 NR structure and orientation shown in Fig. 2B. Simulations performed for 13.5 ns showed an equilibrium conformation that matched the AFM morphology and ∼1 nm height (Fig. 2E). In addition, the relative amino acid flexibility near the N terminus in the simulations (Fig. 2 C and E) and the overlay of the simulated structure on the AFM images indicated that the 0.9 to 1.2 nm periodicity (Fig. 2A) and corresponding bright features at the NR edges (Fig. 2B, Bottom) can be due to pairing of N to N terminus between every two monomers within a NR.
A similar analysis for the p14P2, 14P2Cterm, and p14P2Cterm NRs was limited to six monomers on graphite due to the computationally intensive nature of simulations for the higher number of atoms involved. Despite the limitation, simulations showed that modifying 14P2 with phosphoserine (Fig. 2G) or appending the Cterm domain (SI Appendix, Fig. S6 D and E) generated similar ordered packing in the YINFSY segment of the 14P2 domain on graphite. However, the Cterm domains (DKTKREEVD) in the 14P2Cterm and p14P2Cterm were more disordered than their 14P2 domains and paired up to twist away from the backbone, indicating flexibility at the C-terminal end of the ribbons, consistent with the simulations in solution (Fig. 2D). To further resolve the structure of the Cterm sequences on graphite, we overlaid the simulated β-sheet structure of the 14P2Cterm in solution onto the high-resolution AFM image (Fig. 2F). The overlay showed an excellent match in the width of the β-sheet and the gap between two adjacent NRs, both of which were larger than those for 14P2 NRs (Fig. 2B).
In addition to showing that the most stable predicted structures match that of the single β-sheet structure deduced from AFM and XRD measurements, the MD simulations also predicted that for all peptides investigated, the phenyl residues are bound to the graphite surface by π–π interactions (Fig. 2 E and G, Side View, and SI Appendix, Fig. S6), whereas nearly all hydrophilic side chains, including phosphoserine, glutamic acid, and asparagine, protrude into the solution with a 0.43 to 0.52 nm periodicity that transverses the long axis of the NR (highlighted by dashed yellow circles in Fig. 2 E and G). While detailed in situ physicochemical studies on R2 NRs on HOPG at the molecular level to confirm these predictions are currently not possible, from the simulations and their comparison to the AFM and XRD results, we again conclude that the R2 NRs formed on HOPG are single β-sheets, which are precursors to the cross-β-sheets both formed in bulk solution and observed in vivo, and they present a similar (hydrophilic) NR surface to the solution. Thus, they provide a suitable NR–solution interface for AFM-based nucleation studies.
Calcium Phosphate Nucleation, Growth, and Phase Transformation.
To quantify the impact of Amel NR–solution interfaces on calcium phosphate nucleation rates, we used in situ AFM to investigate nucleation using substrates with R2 NRs, identical to those in Fig. 1 D–H, at pH 7.4 and 25 °C for a range of supersaturations σ (Table 2) over timescales for which nucleation was absent both in solution and on bare HOPG (SI Appendix, Fig. S7). The value of σ was calculated using an equilibrium solubility determined in 2 ways: 1) from the value of the equilibrium constant Ksp reported previously at a different ionic strength (32), and 2) using the ionic activity product at which the postnucleation growth rate extrapolated to zero (SI Appendix, Method 5).
Table 2.
Ion activities and supersaturations with respect to ACP for the three sets of solutions used for mineralization
| Activity (mM) at pH 7.4 | σACP (Ksp = 1.155 × 10−15 M5 from growth rates) | σACP (Ksp = 8.03 × 10−17 M5 from Habraken et al. [32]) | |
|---|---|---|---|
| Ca2+ | HPO42− | ||
| 0.2826 | 3.526 | 0.221 | 0.75 |
| 0.2618 | 3.227 | 0.138 | 0.67 |
| 0.239 | 2.916 | 0.04 | 0.57 |
Time-lapse images revealed that for all values of σ explored here, all five sequences nucleated similarly shaped spherical cap-shaped calcium phosphate particles (Fig. 3 A–E), though at different rates (Fig. 3F). Additional postnucleation characterization by in situ AFM, supported by transmission electron microscopy, revealed that ACP was the first phase to form in all cases (SI Appendix, Figs. S8–S11 and Methods 7–9). These ACP particles grew in size before transforming to fiber- or plate-shaped crystals (Fig. 3 A–E, Panel 5) with sequence-specific growth rates (Fig. 3G) and lifetimes of 14P2Cterm: ∼39 min < p14P2Cterm: 56.63 min < 14P2: 56.71 min < rH174: 125.71 min < p14P2: ∼218.9 min (SI Appendix, Fig. S11). The analysis of the radially integrated electron diffraction data of the initially formed particles confirmed the absence of 0.25 to 0.33 nm d-spacing corresponding to octacalcium phosphate (OCP) or AP found in late-stage mineral (SI Appendix, Fig. S10). The results obtained here are consistent with the identity and dynamics of nuclei formed under identical experimental conditions—with or without proteins—previously characterized using multiple techniques (6, 32, 33). The composition of ACP was previously reported to be Ca2(HPO4)3)2− (32).
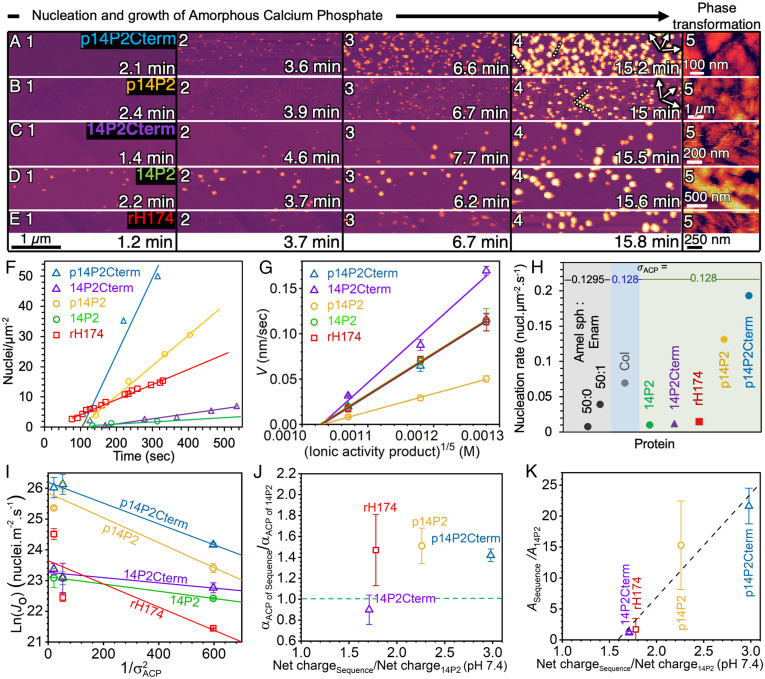
Fig. 3.
Kinetics and energetics of ACP nucleation on peptide and protein NRs. (A–E) Time-lapse of AFM images using constant supersaturation, σACP = 0.221 (σAP = 3.37), at 25 °C and pH 7.4, with t = 0 min defined as the time when solution is introduced into the flow cell. Dotted lines indicate ACP aligned with direction of NRs (arrows). (F) Plot of nuclei number density over time measured for all sequences at σACP = 0.22. Error bar is smaller than data points. (G) Average growth rate of particle height (V) at σACP = 0.04, 0.138, and 0.221. (H) Comparison of nucleation rates for different proteins: Amel nanospheres (Amel sph) coassembled without (50:0) and with (50:1) Enamelin (Enam) at σACP = 0.1295, collagen (Col) at σACP = 0.128, and Amel NRs at σACP = 0.128 (from fits to data in I). (I) Linear fits to Ln(Jo) (nuclei m−2 s−1) measured at different ACP supersaturations (1/σ2ACP) according to Eq. [1]. (J) Ratio of interfacial energy (αACP) and (K) ratio of kinetic prefactor (A) of sequences with respect to 14P2 (αACP of 14P2 = 1.58 mJ m−2, A14P2 = 1.1 × 1010) for each sequence vs. ratio of net charge of sequences with respect to 14P2 (−1.4) at pH 7.4. Dashed line in J shows location of αACP of 14P2 and in K shows linear fit. In all cases, the values of σACP used here are based on the activity product at which growth rates extrapolate to zero. See SI Appendix, Table S6 for αACP calculated based on the Ksp from a previous study (32) and statistics section of Materials and Methods for error analysis and n.
Analysis of the early stage of mineralization on all sequences showed that nucleation rates and nuclei number density were highest on the phosphorylated peptides, p14P2 and p14P2Cterm (Fig. 3F and SI Appendix, Fig. S12), which had particles concentrated along the length of the NRs (Movie S6). In contrast, ACP growth rates were higher on nonphosphorylated peptides than on their phosphorylated versions (Fig. 3G). Furthermore, appending the C terminus to the 14P2 and p14P2 sequences resulted in an increase of 1.3 to 2 times in nucleation rates and a factor of 1.5 to 2.3 in (vertical) growth rates. A comparison to previous results using conformationally dissimilar Amel nanospheres 8 to 9 nm in size (Amel sph) and Amel sph coassembled with enamelin (Enam), a highly charged protein also associated with enamel formation, showed that all NR sequences drive higher nucleation rates than do nanospheres and that the phosphorylated NRs outperform the mixed Amel sph–Enam system, even at lower supersaturations and for the optimal Amel sph:Enam ratio (Fig. 3H, Amel sph: Enam, 50:0 and 50:1) (6). Interestingly, phosphorylated NRs induced ACP nucleation rates 1.9 to 2.8 times larger than observed even on bovine Type I collagen protein (Fig. 3H, Col. σACP = 0.128) (32), which is also rich in proline and phosphoserines and has an extended acicular morphology. These findings show that both sequence and conformation strongly impact template activity and that phosphorylated Amel NRs are the most potent ACP nucleators investigated to date.
Energetics of NR Interface.
To determine the mechanism and underlying energetic factors through which Amel NRs drive ACP nucleation, the data on nucleation rates vs. σ were analyzed using classical nucleation theory (CNT), which has been used previously to analyze heterogeneous nucleation on organic templates (6, 32, 34–36) and has been shown to effectively describe ACP nucleation kinetics (32). CNT predicts that the heterogeneous nucleation rate (Jo) varies exponentially with the effective interfacial energy (αACP) and σACP according to:
| [1] |
| [2] |
where A is the kinetic prefactor, which depends on the density of possible nucleation sites (37), attachment rates, and barriers to ion desolvation (38); ω is the volume of the ACP growth unit, k is Boltzmann’s constant, and T is the absolute temperature (SI Appendix, Method 10 for details).
Fitting the data for all values of σACP revealed two key energetic features of Amel NRs (Fig. 3I). First, the values of αACP for all NR sequences are remarkably small when compared to either nucleation on collagen (40 mJ m−2) or in bulk solution (100 to 150 mJ m−2) (32), ranging from either 1.4 to 2.4 mJ m−2 or 12 to 20 mJ m−2, depending on the method used to determine Ksp. Hence, we infer that nucleation is highly favorable on Amel NRs due to the low interfacial energy. The data also show that both the addition of the C term domain to 14P2 and the phosphorylation of the analogs have minor effects on αACP, with the induced change ranging from a factor of 0.9 to 1.5 (Fig. 3J). The second key feature, which stands in stark contrast to the uniformity of αACP, is a 15- to 19-fold increase in the kinetic prefactor A with phosphorylation (Fig. 3K). Comparison of the relative values of A with the relative increase in the net charge of the peptide revealed a positive linear scaling, likely reflecting higher rates of Ca ion binding or lifetimes of bound Ca ions at the NR–solution interface.
Structure–Function Relationship.
From the perspective of chemical kinetics, the presence of phosphate groups on surfaces should lead to strong binding of multi-ion Ca complexes and a larger kinetic prefactor (36). However, this does not explain why phosphorylated 14P2 NRs have low interfacial energies. Therefore, we investigated structural contributions from the template by comparing the geometric arrangement of the NR charged side chains with the dimer structure of the Ca(HPO4)34− multi-ion complexes shown previously to be the incipient species leading to the formation of ACP through the binding of an additional Ca2+ ion as well as to the formation of OCP and AP (32) through further ion binding reactions.
The Ca–Ca distance in the (Ca2(HPO4)3)2− dimer is approximately 1 nm, and the overall size of the dimer is 1.2 ± 0.2 nm. The superposition of Ca2+ ions in this dimer onto the predicted p14P2 R2 NR conformation shows an excellent match with the spacing of the periodic hydrophilic residues that span the long axis of the NR (Fig. 4A). The dimer could form either over 3 to 4 protein monomers at the phosphoserine sites or over a single protein monomer between phosphoserine and glutamic acid. Thus, the binding of Ca2+ ions to the periodically repeating phosphoserine residues may provide a means to stabilize the dimers of calcium triphosphate and promote the formation of ACP by enhancing kinetics without a drastic impact on interfacial energy (αACP). Interestingly, the protonated a (010) and b (110) facets of OCP and AP have d-spacings of 0.92 nm (or a/2 = b/2 = 0.47 nm), which coincides with the periodicity of hydrophilic residues in Amel NR (Fig. 2A). Therefore, when ACP transforms, the NR may continue to act as a template for OCP or AP due to the structural matching, consistent with both recent studies showing that negatively charged residues likely remain bound to AP phases (39) and the Amel NR model proposed in Carneiro et al. (9), wherein every alternate phosphoserine is available for interaction with the mineral.
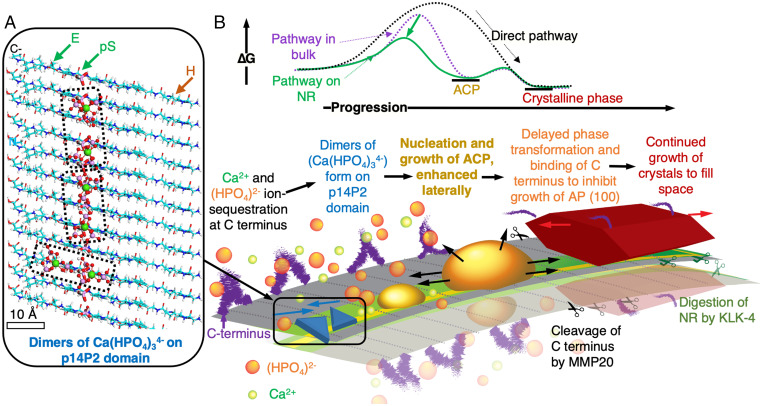
Fig. 4.
Proposed mechanism for full-length Amel NR-guided mineralization. (A) Cartoon of prenucleation clusters (Ca2(HPO4)3)4− superimposed on β-sheet conformation of p14P2 NR shows potential sites for Ca2+ ion docking to allow formation of (Ca2(HPO4)3)2− dimers with 1 nm Ca–Ca distance; negative: glutamic acid (E) and phosphoserine (pS); positive: histidine (H). (B) Proposed role of phosphorylated, cross-β-sheet full-length Amel NR based on energetics and kinetics; NR lowers the barrier (ΔG) for nucleation of ACP by stabilizing dimers of (Ca(HPO4)34−) on 14P2 domain and following a multistep crystallization pathway. Phosphorylated 14P2 domain induces ACP to nucleate and spread laterally. ACP eventually transforms to a crystalline phase (OCP or AP), along with binding of C terminus cleaved by MMP20. The template structure is retained, the crystal continues to grow along the c-axis, and NR is finally enzymatically digested by Kallikrein-4 (KLK-4) (58). In presence of ion-rich PILP, Amel NRs may promote binding of PILP to further enhance the local supersaturation and ACP nucleation rates along the NR long axis.
The experiments performed here differ from a number of calcium phosphate mineralization studies, which utilized a polymer-induced liquid precursor (PILP) phase produced by the addition of acidic macromolecules, such as polyaspartic acid (40–42), including a study that demonstrated that synthetic NRs can template the growth of AP filaments (13). Our results do not rule out the involvement of PILP during ACP nucleation in vivo. However, the stereochemical relationship between Amel NR and calcium phosphate ion complexes, the low interfacial energies, and the enhanced ion binding kinetics are expected to promote ACP nucleation, regardless of whether the ions are delivered from an ion-rich PILP droplet or a less-concentrated calcium phosphate solution like that used in our study. As concluded in these previous studies, PILP likely provides a means for the liquid-phase infiltration of ions into the protein scaffold, but the nucleation of a solid ACP phase is still promoted by charged residues on the scaffold or the complexation of PILP molecules with the scaffold (41). The low energy interface presented by Amel NR likely promotes the binding of the PILP liquid phase, which then creates a high local supersaturation due to the high concentration of calcium and phosphate ions around the acidic macromolecules (43). Because higher supersaturation leads to a higher nucleation rate (Eq. [1] and Fig. 3I), nucleation rates can be dramatically higher with PILP. Thus, wetting by PILP may lead to the rapid formation of ACP along the length of the NR following the energetics presented here, while the transformation of the ACP, guided by the Ca-binding sites, may then result in a single crystal AP filament.
Conclusions
The above experimental findings show that the amyloid structure of Amel NRs, especially the evolutionarily conserved 14P2 self-assembly domain (9, 44), drastically lowers the thermodynamic barrier for ACP nucleation. The atomistic-scale simulations highlight that Amel NR presents a periodic template of hydrophilic residues for mineral formation—without the involvement of non-Amel proteins—that correlates with our experimental results (Fig. 4B). The presence of periodic phosphoserines in NRs dramatically enhances the kinetic prefactor, likely through enhanced Ca2+ ion binding at charged residues, whereas the addition of the hydrophilic Amel C terminus domain has little impact on these two factors, suggesting that amyloid-like Amel NR can retain its structure and mineralizing function even after cleavage of the C terminus domain by MMP20. Whether the C terminus domain then independently acts to inhibit the growth of AP (100) or (010) facets as previously proposed (22, 45), complexes with mineralizing ions to form PILP-like droplets used in previous NR mineralization experiments to produce aligned AP filaments (13), or both, remains to be explored.
The results also provide evidence that one domain of an intrinsically disordered matrix protein can drive self-assembly into a template that stabilizes a mineral phase while leaving another domain flexible to interact with nonmatrix proteins, ions, or crystals (46, 47). The use of such domain-specific interfacial behavior toward crystal nucleation along with careful control of supersaturations can, in principle, be expanded beyond proteins to understanding and engineering crystallization on block copolymer, peptoid, and other amphiphilic polymer templates (48–50).
Beyond fundamentals, the link between the self-assembled molecular structure of Amel NR templates and mineral nucleation points toward a range of translational opportunities. The ability to readily assemble peptides with control over orientation on crystalline surfaces like graphene (51), mica (52), or MoS2 (30), or to array proteins on surfaces patterned at the nanoscale using block copolymer films (53), as well as nanolithography (54), provides a potential path to organizing Amel NRs for engineering mineralized tissue and the synthesis of hybrid materials. Alternatively, the periodic organization of nucleation sites on the NRs suggests a means for creating patterns of quantum dots or oriented crystals of other materials by tailoring the site chemistry. More broadly, achieving predictive hierarchical growth of inorganic crystalline arrays directed by an organic scaffold, inspired by nature, is a long-sought-after vision of bioinspired material science. The robust hierarchical design of amyloid NRs arrayed with modular functional groups around a self-assembling domain provides a general scaffold design upon which that vision may be realized.
Materials and Methods
Synthesis of Peptide Analogs and rH174.
The peptides GHPGYINFSYEVLT (14P2), GHPGYINF(pS)YEVLT (p14P2), GHPGYINFSYEVLTDKTKREEVD (14P2Cterm), and GHPGYINF(pS)YEVLTDKTKREEVD (p14P2Cterm) were commercially synthesized, purified with HPLC (95% purity), and lyophilized at Elim Biopharma Inc. (Hayward, CA). Recombinant human Amel (rH174) was expressed in Escherichia coli BL21 (DE3) and purified according to a previously reported protocol (55). This rH174 lacks the first amino acid residue (Met1) and phosphorylation at Ser16 compared to native human Amel.
Mineral-Free Self-Assembly of NRs.
All solutions were prepared in polypropylene microcentrifuge tubes (Eppendorf and Ted Pella, Inc, USA). Lyophilized peptide or protein was dissolved in 10 mM HCl pH 1.94 (37 wt.% HCl in H2O, 99.999% trace metals basis, Sigma-Aldrich CAS:7647–01-0 diluted with ultrapure deionized water purified by MilliQ Elix Essential 3 and Advantage A10). Solutions were vortexed and sonicated for 15 to 30 min in a Branson ultrasonic bath (M5800H, 40 kHz frequency) until a clear stock solution was obtained. For all AFM experiments, the stock solutions were 1 mg/mL in concentration and molarities of 626.2 μM for 14P2, 596 μM for p14P2, 370.75 μM for 14P2Cterm, 360 μM for p14P2Cterm, and 50.08 μM for rH174. Solutions were further diluted immediately or aged and then diluted based on their application described below in substrates for AFM-based nucleation and high-resolution AFM and SI Appendix. For more details on protocol design, see SI Appendix, Method 11.
Substrates for AFM-based nucleation measurements.
First, 1 mg/mL stock solutions were vortexed and used after 48 h and up to 336 h of incubation at room temperature (25 °C). Aliquots of 0.1 mg/mL were then diluted from the stock using 10 mM HCl (pH 1.94). Next, 50 μL 0.1 mg/mL solution was drop-cast on freshly cleaved HOPG ZYB quality (Mosaic Spread 1.2° ± 0.2°, from Ted Pella or Bruker) and incubated at 37 °C in a sealed chamber (∼100% relative humdity [RH]) for 12 h for consistency with all samples. After incubation, solutions on HOPG were exchanged with 1 mM HCl (pH 3.1) first and then H2O, 3 times each, to remove unbound protein/peptide without disassembly.
High-resolution AFM.
Dilute aliquots were prepared using freshly dissolved 1 mg/mL stock solutions and immediately used. Concentration of 0.01 mg/mL was used for imaging R1-type NRs of 14P2 and other peptide sequences (Fig. 1 I and K and SI Appendix, Figs. S1 and S2), while 0.05 mg/mL was used for R2-type NRs of 14P2 (Figs. 1J and 2B) and 14P2Cterm (Fig. 2F).
In Situ AFM.
Detailed methods for AFM image processing, NR height and width measurements, nucleation rate, and density analysis are provided in SI Appendix, Method 1.
For high-resolution AFM.
For peptides (Fig. 1 I–K), silicon nitride tip cantilevers (Bruker; OTR4, spring constant k: 0.08 N/m or OTR8, spring constant k: 0.15 N/m) were plasma treated for 1 min before use to make them hydrophilic. Substrates were placed on the AFM stage (Cypher VRS, Asylum Research) and the surface was imaged in liquid at room temperature (25 °C). Images of 14P2 R1 NRs (Fig. 1 I and K) were obtained by drop-casting the 0.01 mg/mL onto the freshly cleaved HOPG (ZYB quality, Ted Pella or Bruker) surface and immediately imaged using the tapping mode. A similar process was used for images of R2 NRs with 0.05 mg/mL (Figs. 1J, 2 B and F) by using contact mode.
Self-assembled films on HOPG.
For peptides (Fig. 1 D–H), silicon nitride cantilevers with a silicon tip (Applied Nanostructures HYDRA4V-100NG, spring constant k: 0.088 N/m or Bruker SNL-10, spring constant k: 0.24 N/m or 0.12 N/m) were treated with ultraviolet (UV)/ozone for 15 min. Substrates were placed on the AFM stage (MultiMode 8, Bruker) and operated in the tapping mode, and the surface was imaged in liquid at room temperature (25 °C). For imaging rH174 (Fig. 1H and Inset), silicon tip cantilevers (Arrow UHF, NanoWorld) were treated with UV/ozone for 15 min prior to performing the high-speed tapping mode (5 to 10 Hz scan rate) on the Cypher VRS (Asylum Research).
For nucleation and growth with supersaturated solutions.
Nucleation experiments for all sequences were performed after analysis of the self-assembled NR films using the above method for peptides on the MultiMode8 (Bruker). The protocol for mineralization experiments at constant composition (supersaturation) and analyses are identical to the procedures used for collagen- and Amel nanosphere-functionalized mica in previous publications (6, 32). Three sets of calcium and phosphate solutions at pH 7.4, 2 mM CaCl2, and 19 mM KH2PO4 (σACP = 0.221); 1.75 mM CaCl2 and 16.95 mM KH2PO4 (σACP = 0.138); and 1.5 mM CaCl2 and 14.9 mM KH2PO4 (σAP = 0.04) before mixing were calculated using activities from Visual MINTEQ and experimentally obtained Ksp as well as previously reported Ksp (32) while maintaining a log (activity of Ca2+/activity of PO43-) of 3.87 (Table 2 and SI Appendix, Method 5 and Tables S1–S3). All solutions were independently prepared and filtered 3 times with a cellulose acetate filter (pore size 0.22 μm) before immediate use. The filtered CaCl2 and KH2PO4 solutions were independently and continuously pumped at 25 μL/min for σAP = 0.221, at 31.25 μL/min for σAP = 0.138, and at 37.25 μL/min for σAP = 0.04 and combined at the inlet of the AFM liquid cell using a custom T-junction. The flow rates were experimentally optimized to avoid diffusion-limited nucleation, and solutions were buffered by a high phosphate concentration for at least 3 h after mixing without bulk solution–formed precipitates. The scan size, rate, and sample lines/frame were optimized with several experiments for each sequence and supersaturation to optimize the imaging time and image quality.
Net Charge of Monomers at pH 7.4.
The net charge for each sequence was calculated (SI Appendix, Tables S5 and S6) using the bioinformatics toolbox, Prot pi, available at https://www.protpi.ch/Calculator/ProteinTool. The net charge relative to 14P2 reflects the net charge of sequence/net charge of 14P2.
Molecular Dynamics Simulations.
Simulation models.
Materials Studio (BIOVIA) was utilized to build all-atom models of the peptides and HOPG surfaces. Peptide models were built using the morphology and structural conformation from the AFM and XRD data of Amel NRs assembled at pH 1.94 for 14P2, p14P2, 14P2Cterm, and p14P2Cterm. The entire systems remained charge-neutral by the addition of chloride anions. Virtual π electrons were included for graphite C atoms and C and N atoms in aromatic rings of peptide residues—tyrosine, histidine, and phenylalanine side chains—to precisely describe the features of π electron clouds (SI Appendix, Fig. S14) (31). The IFF parameters for HOPG and virtual π electrons were used as developed earlier using the energy expressions of CHARMM36 and the consistent valence force field (CVFF) for compatibility, which accurately reproduce lattice parameters, surface energies, and hydration energies, following the principle of thermodynamic consistency (29, 31). The force field parameters for the phosphate species were from IFF and the force field parameters for the peptides were from CHARMM36 (56). Further details are given in SI Appendix, Method 4.1.
Peptide assembly in bulk solution.
First, pristine β-sheets (consisting of 6 peptide strands) of all peptide sequences—14P2, p14P2, 14P2Cterm, and p14P2Cterm—with a backbone distance of 0.47 nm were simulated in water at pH 1.94 without HOPG for 13 to 15 ns to reach the equilibrium state (Fig. 2 C and D and SI Appendix, Fig. S5). The total energies of the systems at equilibrium states were then corrected to the target temperature of 298.15 K by utilizing the heat capacity of each system. For a comparative analysis of energy and stability over time for β-sheets vs. other conformations, random coil structures of all sequences were also simulated at the same conditions. Simulation settings and observations are described in SI Appendix, Method 4.2 and 4.3.
Adsorption energy of peptides on HOPG.
Second, the conformations and adsorption energies () of fully extended single-strand peptides on HOPG (SI Appendix, Fig. S6A) were analyzed using a 2-box method (57). Two systems with the same number of atoms and a difference only in peptide position were established for each peptide: the surface system and the away system. The simulations were carried out for 8 ns to reach equilibrium. Simulations of each system were repeated 3 times to obtain average total energies, which were corrected to the target temperature of 298.15 K by utilizing the heat capacity. The was calculated using the following equation:
| [3] |
where is the adsorption energy, is the total energy of the away system, and represents the total energy of the surface system. Details are described in SI Appendix, Method 4.4.
Assembly and stability of peptide β-sheets on HOPG.
Finally, conformations of β-sheets for all sequences were analyzed on the surface of HOPG to investigate the assembly and stability of NRs. The β-sheets were placed on the HOPG (0001) facet with an angle of 30° from the HOPG directions prior to simulation. Models of the 14P2 β-sheet containing 21 peptide strands (box size 73.8000 × 89.4768 × 50.0000 Å3) were simulated on the HOPG surface for 13.5 ns (Fig. 2E). Models of β-sheets for all sequences containing 6 peptide strands (SI Appendix, Fig. S6 B–E) were simulated on the HOPG surface for ∼20 ns and are discussed further in SI Appendix, Method 4.5.
Statistics.
Growth velocity.
Average particle height was measured for n particles that had minimum number of neighboring particles. The error bar in Fig. 3G is the SE (range). For σACP = 0.04: 14P2 n = 5; p14P2 n = 6; 14P2Cterm n = 4; p14P2Cterm n = 4; and rH174 n = 9. For σACP = 0.138: 14P2 n = 10; p14P2 n = 8; 14P2Cterm n = 7; p14P2Cterm n = 6; and rH174 n = 5. For σACP = 0.221: 14P2 n = 7; p14P2 n = 8; 14P2Cterm n = 7; p14P2Cterm n = 6; and rH174 n = 7.
Nucleation rates and surface energy.
Nucleation was repeated at least 5 times on each sequence for each supersaturation (Figs. 3 G and I and SI Appendix, Fig. S11 and Table S4. Data sets with modal slope values, negligible instrumental artifacts, and minimal variations in substrate topography were selected to calculate averages, and error bars represent SD. For σACP = 0.04: 14P2 n = 2; p14P2 n = 2; 14P2Cterm n = 2; p14P2Cterm n = 2; and rH174 n = 2. For σACP = 0.138: 14P2 n = 3; p14P2 n = 3; 14P2Cterm n = 2; p14P2Cterm n = 3; and rH174 n = 2. For σACP = 0.221: 14P2 n= 4; p14P2 n = 2; 14P2Cterm n = 4; p14P2Cterm n = 2; and rH174 n = 2.
Relative interfacial energy and kinetic prefactor.
Error bars represent SE (range) in Fig. 3 J and K and SI Appendix, Tables S5 and S6. The error for kinetic prefactor A was calculated from the error of the intercept ln(A) in Fig. 3I and SI Appendix, Tables S5 and S6, using the following expression: Error of A = (eIntercept × Error of intercept from fitting). Errors of the ratios for each modified sequence reflect the error range of the modified sequence and that of 14P2.
Supplementary Material
Acknowledgments
Experiments were performed at the Pacific Northwest National Laboratory operated by Battelle for the US Department of Energy (DOE); the Environmental Molecular Sciences Laboratory (grid.436923.9), a DOE Office of Science user facility; and Beamline 8.3.1 of the Advanced Light Source, a DOE Office of Science user facility. Authors also acknowledge support from Dr. Paul Ashby at the Molecular Foundry. Funding: Supported by NIH/NIDCR grant R01-DE025709, NSFOAC-1931587, and NSFCMMI-1940335. Simulations were performed using the Argonne Leadership Computing Facility, a DOE Office of Science user facility supported under contract DE-AC02–06CH11357; and the Summit supercomputer, a joint effort of the University of Colorado Boulder and Colorado State University, supported by NSF ACI-1532235 and NSF ACI-1532236.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106965119/-/DCSupplemental.
Data Availability
All data are included in the article and/or SI Appendix.
References
- 1.Smith C. E., Hu Y., Hu J. C.-C., Simmer J. P., Ultrastructure of early amelogenesis in wild-type, Amelx-/-, and Enam-/- mice: Enamel ribbon initiation on dentin mineral and ribbon orientation by ameloblasts. Mol. Genet. Genomic Med. 4, 662–683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beniash E., Metzler R. A., Lam R. S. K., Gilbert P. U. P. A., Transient amorphous calcium phosphate in forming enamel. J. Struct. Biol. 166, 133–143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandya M., et al. , Posttranslational amelogenin processing and changes in matrix assembly during enamel development. Front. Physiol. 8, 790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moradian-Oldak J., Amelogenins: Assembly, processing and control of crystal morphology. Matrix Biol. 20, 293–305 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Fang P.-A., Conway J. F., Margolis H. C., Simmer J. P., Beniash E., Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc. Natl. Acad. Sci. U.S.A. 108, 14097–14102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao J., et al. , Control of calcium phosphate nucleation and transformation through interactions of enamelin and amelogenin exhibits the “Goldilocks Effect”. Cryst. Growth Des. 18, 7391–7400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fincham A. G., et al. , Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J. Struct. Biol. 115, 50–59 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Paine M. L., et al. , Enamel biomineralization defects result from alterations to amelogenin self-assembly. J. Struct. Biol. 132, 191–200 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Carneiro K. M. M., et al. , Amyloid-like ribbons of amelogenins in enamel mineralization. Sci. Rep. 6, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Avila O., et al. , Self-assembly of filamentous amelogenin requires calcium and phosphate: From dimers via nanoribbons to fibrils. Biomacromolecules 13, 3494–3502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pautard F. G. E., An x-ray diffraction pattern from human enamel matrix. Arch. Oral Biol. 3, 217–220 (1961). [DOI] [PubMed] [Google Scholar]
- 12.Jodaikin A., Traub W., Weiner S., Protein conformation in rat tooth enamel. Arch. Oral Biol. 31, 685–689 (1986). [DOI] [PubMed] [Google Scholar]
- 13.Bai Y., et al. , Protein nanoribbons template enamel mineralization. Proc. Natl. Acad. Sci. U.S.A. 117, 19201–19208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y., et al. , A N-terminus domain determines amelogenin’s stability to guide the development of mouse enamel matrix. J. Bone Miner. Res. 36, 1781–1795 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai P., Warshawsky H., Morphological studies on the distribution of enamel matrix proteins using routine electron microscopy and freeze-fracture replicas in the rat incisor. Anat. Rec. 212, 1–16 (1985). [DOI] [PubMed] [Google Scholar]
- 16.Travis D. F., Glimcher M. J., The structure and organization of, and the relationship between the organic matrix and the inorganic crystals of embryonic bovine enamel. J. Cell Biol. 23, 447–497 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rönnholm E., An electron microscopic study of the amelogenesis in human teeth. I. The fine structure of the ameloblasts. J. Ultrastruct. Res. 6, 229–248 (1962). [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Avila O. M., et al. , Self-assembly of amelogenin proteins at the water-oil interface. Eur. J. Oral Sci. 119 (suppl. 1), 75–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanii B., Martinez-Avila O., Simpliciano C., Zuckermann R. N., Habelitz S., Matching 4.7-Å XRD spacing in amelogenin nanoribbons and enamel matrix. J. Dent. Res. 93, 918–922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelberth S. A., et al. , Progression of self-assembly of amelogenin protein supramolecular structures in simulated enamel fluid. Biomacromolecules 19, 3917–3924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin N. Y., et al. , Amelogenin phosphorylation regulates tooth enamel formation by stabilizing a transient amorphous mineral precursor. J. Biol. Chem. 295, 1943–1959 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friddle R. W., et al. , Single-molecule determination of the face-specific adsorption of amelogenin’s C-terminus on hydroxyapatite. Angew. Chem. Int. Ed. Engl. 50, 7541–7545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gungormus M., et al. , Cementomimetics—constructing a cementum-like biomineralized microlayer via amelogenin-derived peptides. Int. J. Oral Sci. 4, 69–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao J., et al. , The energetic basis for hydroxyapatite mineralization by amelogenin variants provides insights into the origin of amelogenesis imperfecta. Proc. Natl. Acad. Sci. U.S.A. 116, 13867–13872 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowalewski T., Holtzman D. M., In situ atomic force microscopy study of Alzheimer’s beta-amyloid peptide on different substrates: New insights into mechanism of beta-sheet formation. Proc. Natl. Acad. Sci. U.S.A. 96, 3688–3693 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruggeri F. S., et al. , Identification and nanomechanical characterization of the fundamental single-strand protofilaments of amyloid α-synuclein fibrils. Proc. Natl. Acad. Sci. U.S.A. 115, 7230–7235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serpell L. C., Alzheimer’s amyloid fibrils: Structure and assembly. Biochim. Biophys. Acta 1502, 16–30 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Nelson R., et al. , Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinz H., Lin T.-J., Mishra R. K., Emami F. S., Thermodynamically consistent force fields for the assembly of inorganic, organic, and biological nanostructures: The INTERFACE force field. Langmuir 29, 1754–1765 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Chen J., et al. , Building two-dimensional materials one row at a time: Avoiding the nucleation barrier. Science 362, 1135–1139 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Pramanik C., Gissinger J. R., Kumar S., Heinz H., Carbon nanotube dispersion in solvents and polymer solutions: Mechanisms, assembly, and preferences. ACS Nano 11, 12805–12816 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Habraken W. J. E. M., et al. , Ion-association complexes unite classical and non-classical theories for the biomimetic nucleation of calcium phosphate. Nat. Commun. 4, 1–12 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Chien Y.-C., et al. , Using biomimetic polymers in place of noncollagenous proteins to achieve functional remineralization of dentin tissues. ACS Biomater. Sci. Eng. 3, 3469–3479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Q., et al. , The thermodynamics of calcite nucleation at organic interfaces: Classical vs. non-classical pathways. Faraday Discuss. 159, 509–523 (2012). [Google Scholar]
- 35.Giuffre A. J., Hamm L. M., Han N., De Yoreo J. J., Dove P. M., Polysaccharide chemistry regulates kinetics of calcite nucleation through competition of interfacial energies. Proc. Natl. Acad. Sci. U.S.A. 110, 9261–9266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamm L. M., et al. , Reconciling disparate views of template-directed nucleation through measurement of calcite nucleation kinetics and binding energies. Proc. Natl. Acad. Sci. U.S.A. 111, 1304–1309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kashchiev D., van Rosmalen G. M., Review: Nucleation in solutions revisited. Cryst. Res. Technol. 38, 555–574 (2003). [Google Scholar]
- 38.De Yoreo J. J., Vekilov P. G., Principles of crystal nucleation and growth. Rev. Mineral. Geochem. 54, 57–93 (2003). [Google Scholar]
- 39.Hoff S. E., Liu J., Heinz H., Binding mechanism and binding free energy of amino acids and citrate to hydroxyapatite surfaces as a function of crystallographic facet, pH, and electrolytes. J. Colloid Interface Sci. 605, 685–700 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Jee S.-S., Thula T. T., Gower L. B., Development of bone-like composites via the polymer-induced liquid-precursor (PILP) process. Part 1: Influence of polymer molecular weight. Acta Biomater. 6, 3676–3686 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Nudelman F., et al. , The role of collagen in bone apatite formation in the presence of hydroxyapatite nucleation inhibitors. Nat. Mater. 9, 1004–1009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y.-X., et al. , Involvement of prenucleation clusters in calcium phosphate mineralization of collagen. Acta Biomater. 120, 213–223 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Kim Y. Y., Douglas E. P., Gower L. B., Patterning inorganic (CaCO3) thin films via a polymer-induced liquid-precursor process. Langmuir 23, 4862–4870 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Wald T., et al. , Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif. Proc. Natl. Acad. Sci. U.S.A. 114, E1641–E1650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iline-Vul T., et al. , Osteopontin regulates biomimetic calcium phosphate crystallization from disordered mineral layers covering apatite crystallites. Sci. Rep. 10, 15722 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boskey A. L., Villarreal-Ramirez E., Intrinsically disordered proteins and biomineralization. Matrix Biol. 52–54, 43–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao A., et al. , Stabilization of mineral precursors by intrinsically disordered proteins. Adv. Funct. Mater. 28, 1802063 (2018). [Google Scholar]
- 48.Oleske K. W., et al. , Nanopatterning of crystalline transition metal oxides by surface templated nucleation on block copolymer mesostructures. Cryst. Growth Des. 17, 5775–5782 (2017). [Google Scholar]
- 49.Kim Y.-Y., et al. , Hydroxyl-rich macromolecules enable the bio-inspired synthesis of single crystal nanocomposites. Nat. Commun. 10, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan F., et al. , Controlled synthesis of highly-branched plasmonic gold nanoparticles through peptoid engineering. Nat. Commun. 9, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarikaya M., Tamerler C., Jen A. K.-Y., Schulten K., Baneyx F., Molecular biomimetics: Nanotechnology through biology. Nat. Mater. 2, 577–585 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Pyles H., Zhang S., De Yoreo J. J., Baker D., Controlling protein assembly on inorganic crystals through designed protein interfaces. Nature 571, 251–256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stel B., et al. , Contrasting chemistry of block copolymer films controls the dynamics of protein self-assembly at the nanoscale. ACS Nano 13, 4018–4027 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Lee K.-B., Park S.-J., Mirkin C. A., Smith J. C., Mrksich M., Protein nanoarrays generated by dip-pen nanolithography. Science 295, 1702–1705 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Svensson Bonde J., Bulow L., One-step purification of recombinant human amelogenin and use of amelogenin as a fusion partner. PLoS One 7, e33269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang J., MacKerell A. D. Jr., CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz H., Computational screening of biomolecular adsorption and self-assembly on nanoscale surfaces. J. Comput. Chem. 31, 1564–1568 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Lu Y., et al. , Functions of KLK4 and MMP-20 in dental enamel formation. Biol. Chem. 389, 695–700 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article and/or SI Appendix.