Figure 1. Technical detection of pathogenic/likely pathogenic (P/LP) germline variants affecting moderate- to high-penetrance cancer susceptibility genes by tumor-only sequencing.
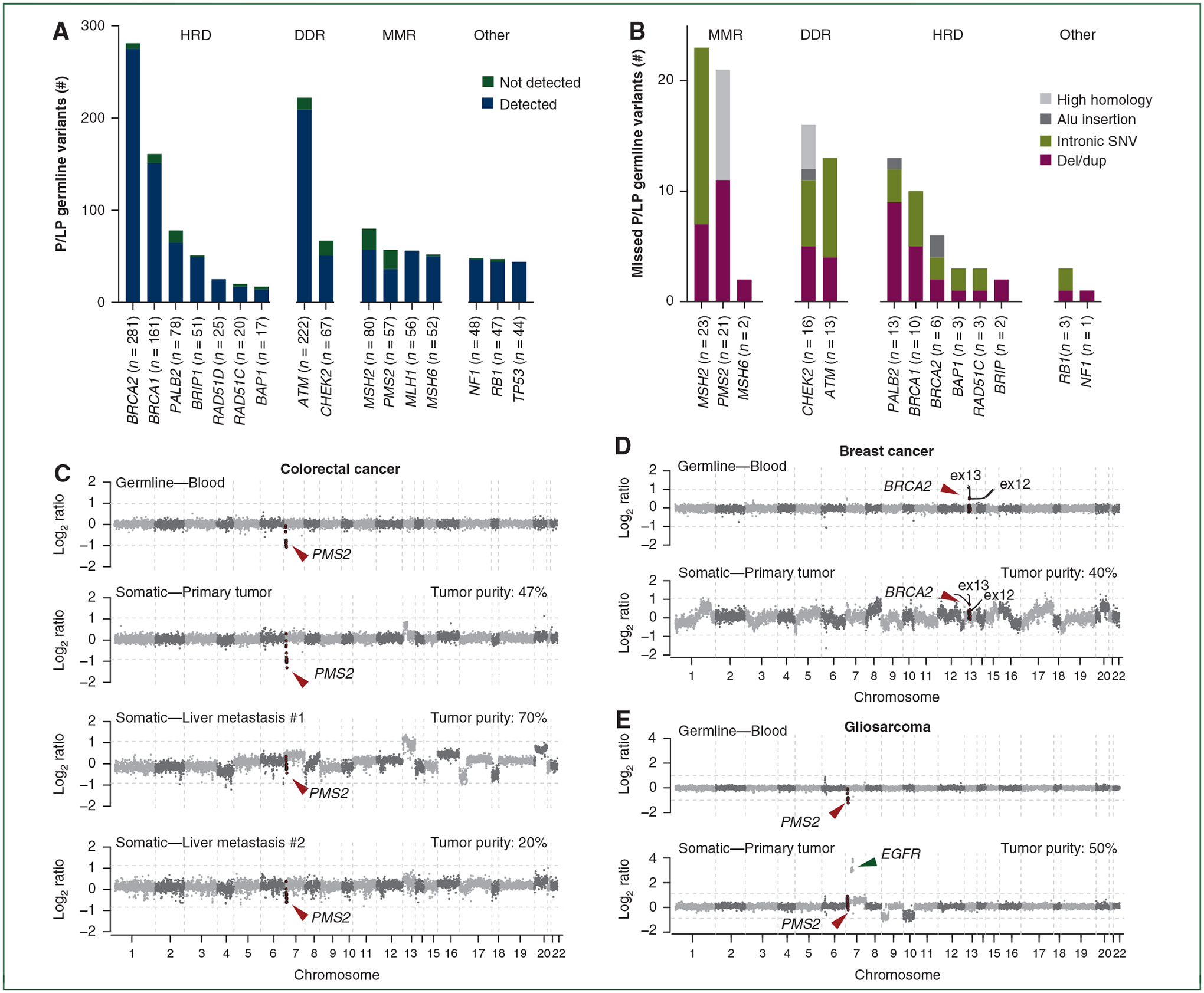
(A) P/LP germline variants technically not detected by tumor-only sequencing by gene and biological process. (B) P/LP germline variants not detected by tumor-only sequencing according to variant type. (C-E) Copy number plots depicting segmented Log2 ratios (y-axis) according to genomic position (x-axis) of blood and tumor samples in (C) a colorectal carcinoma with a PMS2 germline whole gene deletion that is detectable in the primary tumor specimen but obscured due to genomic instability in two liver metastases, (D) a breast cancer with a germline BRCA2 exon 12 and 13 duplication not detected in the tumor specimen due to somatic copy number variants (CNVs) and (E) a gliosarcoma with a germline PMS2 whole gene deletion harboring a somatic EGFR amplification and chromosome 7 gain which prevents the detection of the germline PMS2 deletion.
HRD, homologous recombination deficiency; DDR, DNA damage response; MMR, mismatch repair; SNV, single-nucleotide variant.
