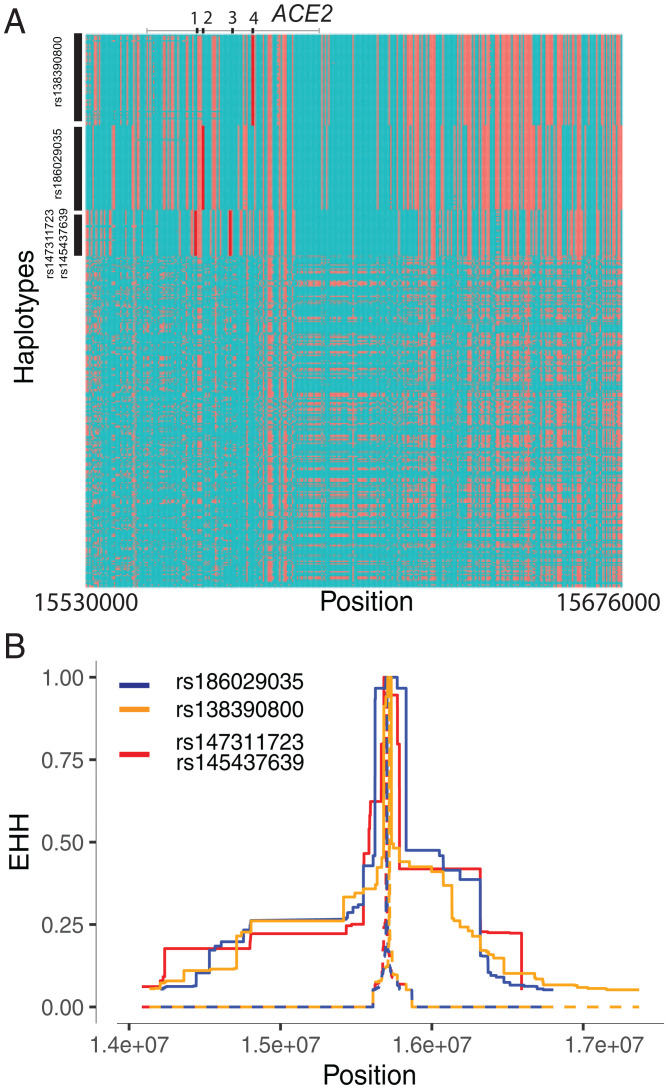
Fig. 2.
Natural selection signatures at ACE2 in the Cameroon CAHG populations. (A) Haplotypes over 150 kb flanking ACE2 in CAHG populations. The x axis denotes the genetic variant position, and the y axis represents haplotypes. Each haplotype (one horizontal line) is composed of the genetic variants (columns). Red dots indicate the derived allele, while green dots indicate the ancestral allele. Haplotypes surrounded by the upper left vertical black lines suggest that these haplotypes carry derived allele(s) of the labeled variant near the corresponding black line. For example, the first black line denotes all the haplotypes that have the derived allele at rs138390800 (dark red line). Haplotypes carrying rs138390800, rs147311723, rs145437639, and rs186029035 show more homozygosity than other haplotypes; 1, 2, 3, and 4 along the top of the plot denote positions for rs147311723, rs186029035, rs145437639, and rs138390800, respectively. (B) EHH of rs138390800, rs186029035, and rs147311723 (rs145437639 is in strong LD with rs147311723) at ACE2 in CAHG populations.