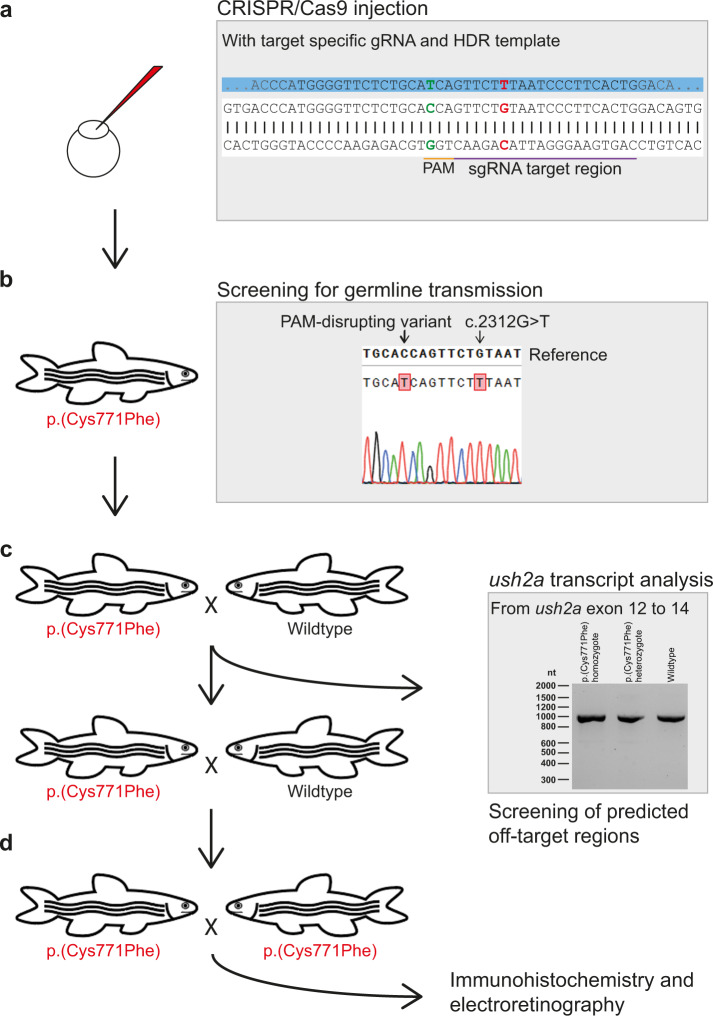
Fig. 2. Generation of the usherinp.(Cys771Phe) zebrafish model.
a Eggs were injected at a one-cell stage with CRISPR/Cas9 mix containing both the target specific single guide RNA (sgRNA) and the 126 nt homology directed repair (HDR) template (partially depicted, in blue) for incorporation of the c.2312 G > T (p.(Cys771Phe)) variant (in red) and protospacer-adjacent motif (PAM, in orange) disrupting variant (c.2304 C > T, in green). The sgRNA target region is depicted in purple. b Once zebrafish were at reproductive age, eggs of the initially injected zebrafish were screened for transmission of the c.2312 G > T and PAM disturbing variant. Three out of ten zebrafish that were screened transmitted the c.2312 G > T variant to their offspring. c Crossbreeding of a c.2312 G > T (p.(Cys771Phe)) zebrafish with a wildtype zebrafish was performed for two generations to reduce unforeseen off-target effects. After the first crossbreeding, genomic DNA was screened for predicted off-target effects of our CRISPR-Cas9 strategy and RNA of homozygous larvae was screened from exon 12 to 14 for deviations on transcript level. d Two p.(Cys771Phe) zebrafish were crossbred with each other to produce homozygous zebrafish. The phenotype of five-day-old larvae was then investigated with immunohistochemistry and electroretinography.