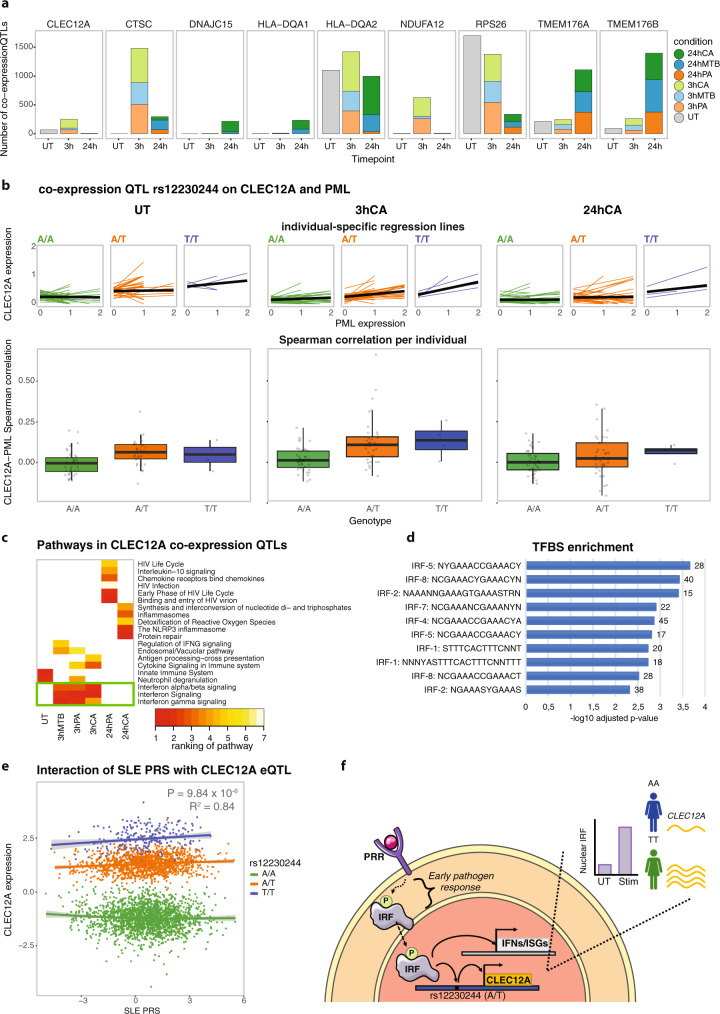
Fig. 4. Interferon regulatory factor affects CLEC12A co-expression QTLs upon 3 h pathogen stimulation in monocytes.
a Number of co-expression QTLs identified in each of the stimulation‒timepoint combinations for those co-expression QTLs with over 100 co-expression QTLs in at least one condition. The 3 h and 24 h timepoint are colored by pathogen stimulation (green: C. albicans (CA), blue: M. tuberculosis (MTB), orange: P. aeruginosa (PA). Co-expression QTL summary statistics can be found in Supplementary Data 11. b The lines in the top plots show co-expression between CLEC12A and PML (most significant co-expression QTL across the 3 h stimulation conditions) for individual cells in the untreated (left), 3 h CA (middle) and 24 h CA (right) condition. In these plots, individual-specific regression lines are shown, split by genotype. The average genotype-specific regression lines are shown in black. The bottom boxplots depict Spearman’s rank correlation between CLEC12A and PML expression, stratified by SNP rs12230244 genotype in the monocytes per individual, in the untreated (left), 3 h CA (middle) and 24 h CA (right) stimulated cells (the V2 chemistry data is plotted). Each data point shows a single individual. Boxplots show median, first and third quartiles, and 1.5× the interquartile range. c Heatmap of the top-5 enriched pathways within the co-expressed CLEC12A co-eQTL genes per stimulation‒timepoint combination. Per combination, pathways are ranked based on significance. White indicates that the pathway was not found to be enriched in that specific stimulation‒timepoint combination. The green box highlights pathways that are associated with all 3 h stimulation conditions. d Top 10 enriched putative transcription factor binding sites within the CLEC12A co-expression QTL genes that: (1) showed a more positive strength of the co-expression relationship in individuals with the TT as opposed to the AA genotype and (2) were identified in the 3 h stimulated (outer join) monocytes using the TRANSFAC database. Enrichment of putative transcription factor binding sites was defined using a g:SCS multiple testing correction method, applying a significance threshold of 0.05. e Co-expression QTL analysis for CLEC12A-SNP rs12230244 against the SLE polygenic risk score (PRS) (calculated using those SLE GWAS SNPs with a P-value threshold of <5 × 10−8) using whole-blood bulk expression data from 3553 individuals (BIOS consortium). A one-tailed F-test (coefficient = 0.04, standard error = 0.01, f-value = 19.60, p-value = 9.84 × 10−6, R2 = 0.84) was used to determine whether the distribution of the squared residuals with the SLE PRS as interaction term was significantly smaller than without. f Proposed mechanism of action of CLEC12A co-expression QTLs. When pathogen-associated molecular patterns bind to a pattern recognition receptor (PRR), a signaling cascade is initiated that eventually results in phosphorylation of interferon regulatory factors (IRFs). Phosphorylated IRF then translocates to the nucleus, where it binds to specific DNA motifs such as IFN-stimulated response elements. This can then activate transcription of IFNs and IFN-stimulated genes (ISGs). Additionally, IRF is expected to bind to a region containing SNP rs12230244 (or any another SNP in high LD), thereby regulating CLEC12A expression. In this case, depending on the SNP genotype, the IRF binding and activation of CLEC12A expression is expected to be stronger (TT genotype) or weaker (AA genotype). Many of the identified CLEC12A co-expression QTL genes are involved in the IFN pathway (see panel b). This has to be the result of a common upstream factor (i.e., IRF) of CLEC12A transcription that can also activate IFNs and ISGs, but cannot be the result of a downstream regulator because this would have led to trans-eQTL effects for the same SNP rs12230244 (which we do not observe). The number of individuals and cells included in each analysis can be found in the Source Data file.