Abstract
During a microbiological and genomic surveillance study conducted to investigate the molecular epidemiology of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from community-acquired urinary tract infections (UTI) and commercial meat samples, in a Brazilian city with a high occurrence of infections by ESBL-producing bacteria, we have identified the presence of CTX-M (-2, -14, -15, -24, -27 and -55)-producing E. coli of international clones ST38, ST117, ST131 and ST354. The ST131 was more prevalent in human samples, and worryingly the high-risk ST131-C1-M27 was identified in human infections for the first time. We also detected CTX-M-55-producing E. coli ST117 from meat samples (i.e., chicken and pork) and human infections. Moreover, the clinically relevant CTX-M-24-positive E. coli ST354 clone was detected for the first time in human samples. In summary, our results highlight a potential of commercialized meat as a reservoir of high-priority E. coli lineages in the community, whereas the identification of E. coli ST131-C1-M27 indicates that novel pandemic clones have emerged in Brazil, constituting a public health issue.
Subject terms: Computational biology and bioinformatics, Microbiology, Molecular biology
Introduction
Escherichia coli is a commensal of the human intestinal tract and most warm-blooded mammals, and an important pathogen for humans and animals1–3. In humans, urinary tract infections (UTIs) are the second most common bacterial infection managed in primary care, where uropathogenic E. coli (UPEC) is responsible for 75% to 95% of the cases1,4. The increasing antimicrobial resistance of UPEC has been of concern, with infections caused by antimicrobial-resistant (AMR) bacteria as extended-spectrum β-lactamase (ESBL)-producing E. coli representing a significant health care issue, since it compromises the effective treatment, being responsible for a large number of morbidity and mortality1,3,5.
Since E. coli can act as a large reservoir of resistance genes that directly impact treatment in human and veterinary medicine, the debate over the transmission of multidrug-resistant E. coli strains between animals and humans, through numerous pathways, has become increasingly discussed6,7. However, the relation between food-producing animals, humans, the environment, and the transmission of these resistant pathogens is not yet fully understood8,9.
Specifically, the isolation of ESBL-producing E. coli from food-production animals has been increasingly reported worldwide, mostly from chicken meat1,2. In this respect, the excessive use of antimicrobials in livestock has promoted the emergence of antimicrobial-resistant pathogens in humans. On the other hand, the consumption of meat, direct contact with colonized animals, or manure spread in the environment have been described as sources for the transmission of AMR pathogens from livestock to humans3,4. Besides, antimicrobial resistance gene transfer may occur between different bacterial species in the gut of animals and humans5.
The CTX-M type is one of the largest groups of ESBL, and recent studies that addressed the epidemiology of these enzymes in Brazil, show that CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-15 are predominant variants in this country6–8. Many types of CTX-M-producing E. coli have been recognized as belonging to specific clones commonly isolated from UTI cases characteristic of a particular region and that are spread throughout the world. Some studies show that CTX-M genotypes of isolates from foods sometimes correspond with locally dominant human clones9,10. In this respect, Escherichia coli sequence type (ST) 131 is a type of pandemic clone associated with ESBL production11,12. Studies describe that the spread of ST131 is stimulated by a specific sub-lineage called clade C or C/H30 and the reason for its successful spread around the world is still not fully understood13. Although studies have shown that CTX-M-15 production is strongly involved with the predominant ST131 C2/H30Rx subgroup, the emerging C1 subgroup is related to other CTX-M enzymes, such as CTX-M-14 and CTX-M-2712,14,15.
Recently, reports of clade C1-M27 in humans, animals and the environment have been increasing in several countries such as Australia, Canada, France, Germany, Italy, Japan, Netherlands, Spain, Thailand and USA8,12,14. The rapid transmission of ST131-C1-M27 becomes a challenge for global health, as it highlights an evolution of ESBLs caused by the ST131 clone and reinforces the complex structure of this clone12,14–16.
Considering the impact of the AMR in Brazil, both in human medicine as in livestock, and the need for understanding this panorama, we conducted a next-generation sequencing (NGS)-based analysis, within a One Health perspective, in order to assess national transmission of CTX-M-producing E. coli isolated from meat products and human patients.
Results
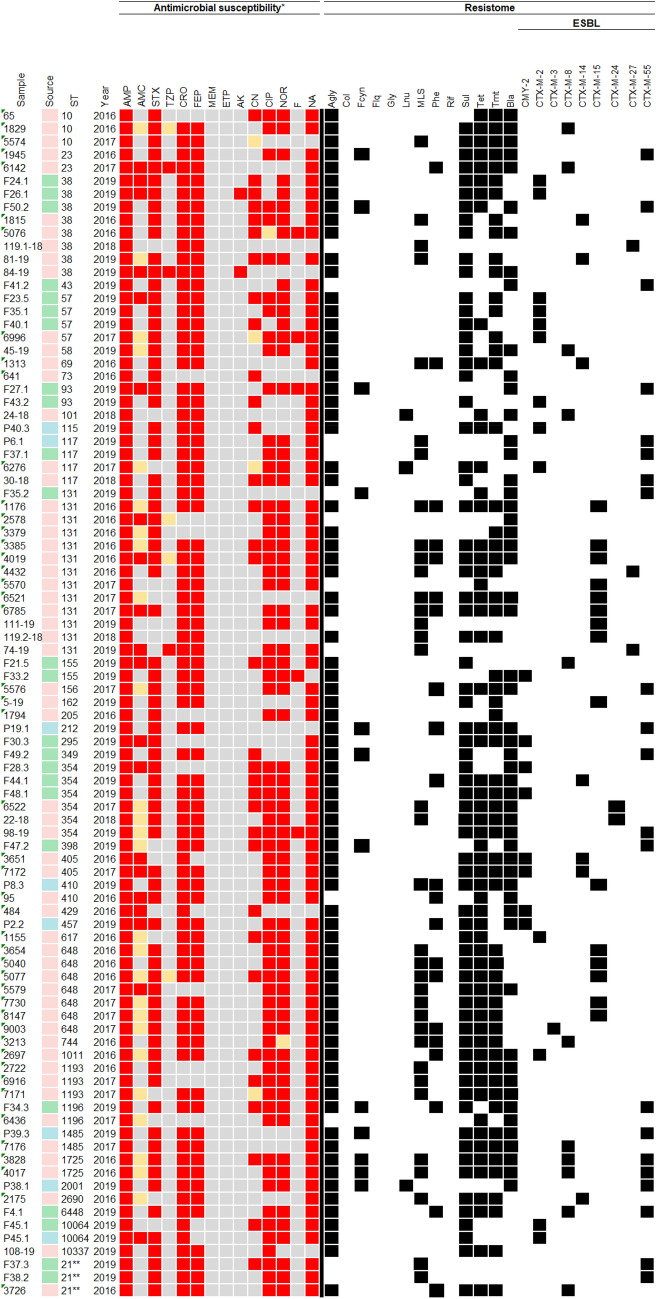
The results about source, ST, antimicrobial susceptibility and resistome for each of 91 E. coli isolates included in this study are quoted in Table 1, more information about the isolates can be seen in "Table 1 complete" in the supplementary material. High rates of resistance to ampicillin (100%), ceftriaxone (87.9%), nalidixic acid (87.9%), cefepime (83.5%), trimethoprim-sulfamethoxazole (82.4%), nitrofurantoin (76.9%), norfloxacin (75.8%) and ciprofloxacin (72.5%) were confirmed, whereas less than half showed resistance to gentamicin (36.3%) and amoxicillin/clavulanate (22%). Only three (3.3%) isolates were resistant to piperacillin-tazobactam and two (2.2%) to amikacin. Some isolates also showed intermediate resistance levels to amoxicillin/clavulanate (28.6%), to piperacillin-tazobactam and gentamicin (4.4%), and to ciprofloxacin and norfloxacin 1.1%).
Table 1.
Heatmap shows sample code, its origin, ST, year of isolation, AMR profile and resistome.
Color legend: pink: human; green: chicken meat; blue: pork; red: resistance; orange: intermediate resistance; gray: susceptibility, black: presence confirmed.
Legend of antimicrobials: AMP ampicillin, AMC amoxicillin-clavulanate, STX trimethoprim-sulfamethoxazole, TZP piperacillin-tazobactam, CRO ceftriaxone, FEP cefepime, MEM meropenem, ERP ertapenem, AK amikacin, CN gentamicin, CIP ciprofloxacin, NOR norfloxacin, F nitrofurantoin, NA nalidixic acid.
Legend of resistome: Agly aminoglycosides, Col colistin, Fcyn fosfomycin, Flq fluoroquinolones, Gly glycopeptides, MSL macrolides, Phe phenicols, Rif rifampin, Sul sulfonamides, Tet tetracyclines, Tmt trimethoprim, Bla β-lactam, Lnu lincosamides.
**Escherichia coli scheme sequencetype II (Institut Pasteur).
Genomic analysis predicted 57 genes associated with resistance to aminoglycosides (n = 15), β-lactams (n = 12), trimethoprim (n = 8), phenicols (n = 5), tetracyclines (n = 4), macrolides (n = 4), sulfonamides (n = 3), quinolones (n = 3), lincosamides (n = 2) and fosfomycin (n = 1). For aminoglycosides, the most prevalent genes were strA and strB (39.6%), followed by the aadA1 (36.3%). Genes dfrA17 and dfrA1 genes, associated with resistance to trimethoprim, were detected in 31 (34.1%) and 13 (14.3%) isolates, respectively. Genes related to phenicols resistance had similar prevalence, being catB3 (8.8%), floR (5.5%), catA1(4.4%) and cmlA1 (4.4%). Concerning to tetracyclines resistance, tet(A) (38.5%) and tet(B) (27.5%) genes were identified. For macrolides resistance, the mph(A) gene (29.7%) was found, whereas sulfonamides resistance sul1 (56%) and sul2 (53.8%) genes were predicted. Genes conferring resistance to lincosamides (lnu-type) and fosfomycin (fosA) were identified in three E. coli strains (3.3%).
Resistance to β-lactams was mediated by blaTEM-1A (1.5%), blaTEM-1B (32.3%), blaOXA-1 (5.3%), blaCMY-2 (6.0%) and blaCTX-M (54.9%). Among ESBL genes eight variants were identified, with predominance of blaCTX-M-55 (25.9%), which was identified in E. coli strains isolated from chicken meat (n = 11) and pork meat (n = 4), and from human hosts (n = 6). The blaCTX-M-15 ESBL gene was predominantly in human isolates (n = 14), being further identified in only one pork isolate. On the other hand, the blaCTX-M-2 gene was also detected in 13 isolates (16%), from chicken meat (n = 7), human (n = 4) and pork meat (n = 2) samples. CTX-M-8 and CTX-M-14 ESBL genes were present in nine and six human isolates, and two and one chicken meat isolates, respectively. CTX-M-24 (n = 2), CTX-M-27 (n = 3) and CTX-M-3 (n = 1) genes were present only in human isolates. The blaCMY-2 gene was predominantly found in chicken meat samples (n = 4), followed by urine samples (n = 3) and pork meat (n = 1).
In this study, 11 types of plasmid replicons were predicted, which belonged to the Col, IncA/C2, IncB/O/K/Z, IncF, IncI, IncQ, IncR, IncY, IncX, p0111 and IncN families. In human isolates, the most frequent were IncI1[ST-113] (n = 9), IncF[F-: A-: B-] (n = 7), IncF[F1: A2: B20] (n = 5), IncF[F48: A1: B49] (n = 5) and p0111 (n = 5). In chicken meat isolates, IncF[F18: A-: B1] (n = 8), p0111 (n = 7) and IncN[unknown ST] (n = 5) were the most frequent Inc-type plasmid identified. In isolates of pork meat, the most frequent incompatibility groups were IncN[unknown ST] (n = 4) and IncF[F33: A-: B1] (n = 3).
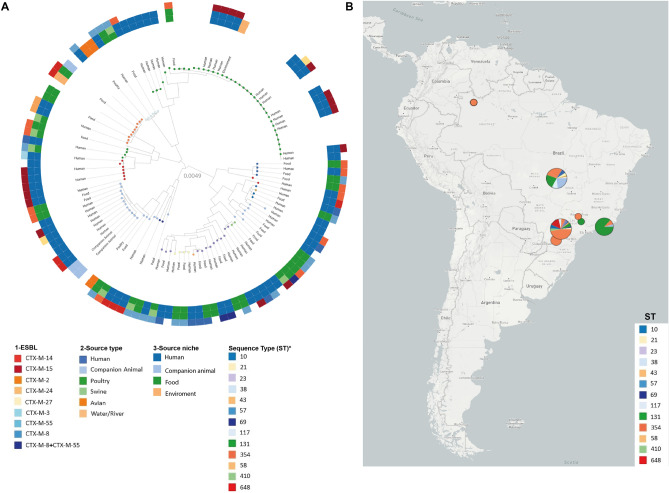
In total, 40 sequence types (STs) were identified, the most prevalent were ST131 (n = 13), ST38 (n = 8), ST648 (n = 7), and ST354 (n = 6). Some STs were detected in strains isolated from more than one source, confirming clonal dissemination between humans and chicken meat. Lineages of ST38, ST131, ST354, and ST1196 were found in both urine and chicken meat. The ST410 was the only observed in urine (n = 1) and pork (n = 1) strains. The ST117 was identified in samples collected from the three sources investigated, with two strains being isolated from urine, and one from chicken meat and pork, respectively. The clonal relatedness among isolates characterized in this study, and their distribution in Brazil, is shown in Fig. 1A,B. Additionally, it is possible to observe that Brazilian isolates of ST131 clustered with isolates from United Kingdom (https://microreact.org/project/2mKg54AHdWj5xdJ5VFejY8). All information and genes detected in this study are quoted in supplementary material (Table 1) (https://doi.org/10.6084/m9.figshare.17108402.v1).
Figure 1.
(A) Escherichia coli Phylogenomic SNP tree with circular heatmap shows (outside-in reading rings): ESBL-gene (first inner ring); source type (second inner circle) and source niches (third inner circle). (B) Distribution map of Escherichia coli sequence type (ST) in Brazilian regions. Asterisk: bubbles colors in phylogenomic branches.
Some important associations were observed, such as one E. coli ST38 isolate producing CTX-M-27 and carrying the IncF[F2:A-:B10] plasmid, and E. coli ST131 producing CTX-M-15 carrying various IncF-type plasmids (i.e., IncF[F2:A-:B1], IncF[F2:A1:B-], IncF[F31:A4:B1], IncF[F36:A4:B1] and IncF[F51:A1:B-]). Other important findings are the identification of CTX-M-27 E. coli ST131 carrying plasmids IncF[F1:A2:B20], and the co-production of CTX-M-8 and CTX-M-55 by E. coli ST1725 and ST6448 harboring IncF[F24:A-:B73], IncF[F33:A-:B1] and/or IncI1[ST-113] plasmids.
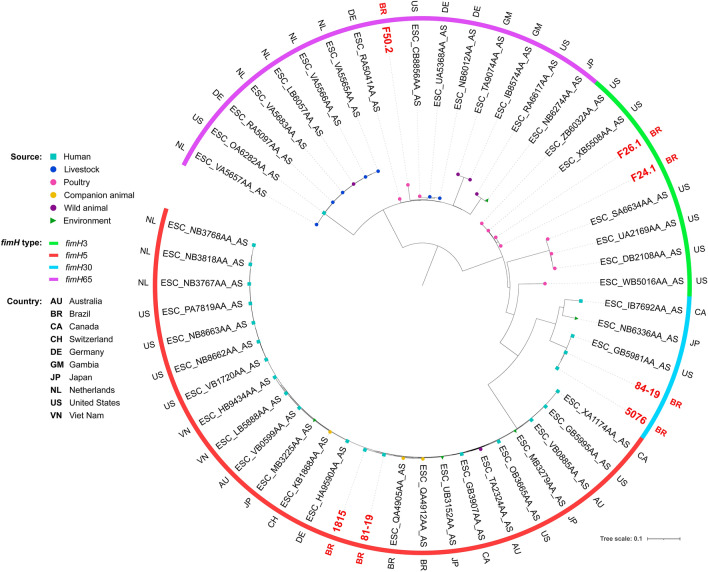
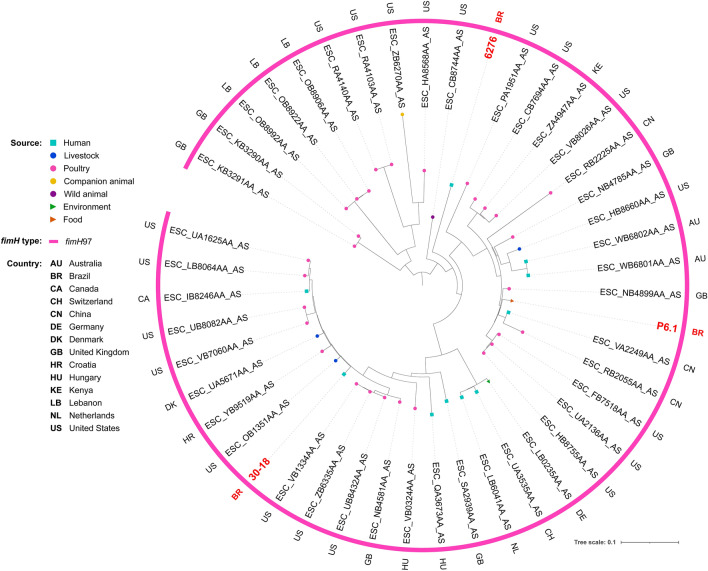
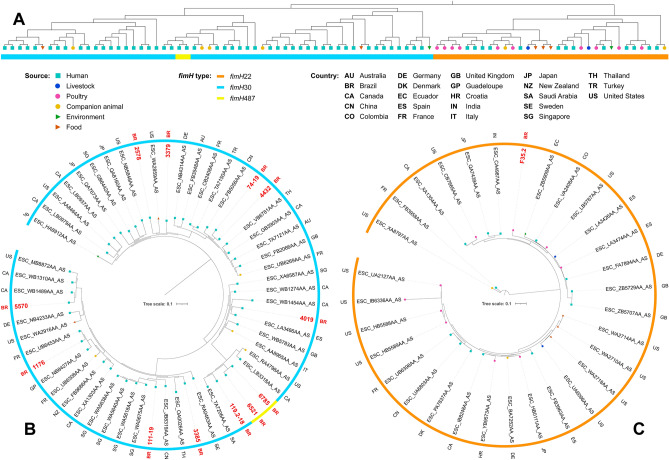
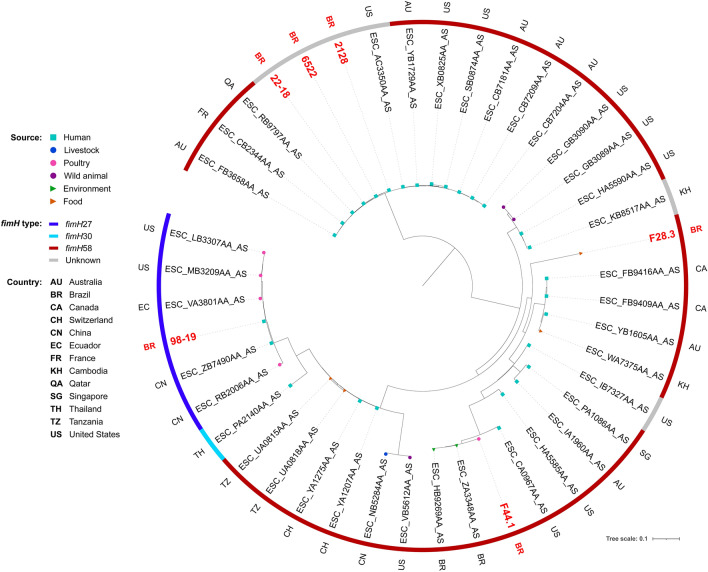
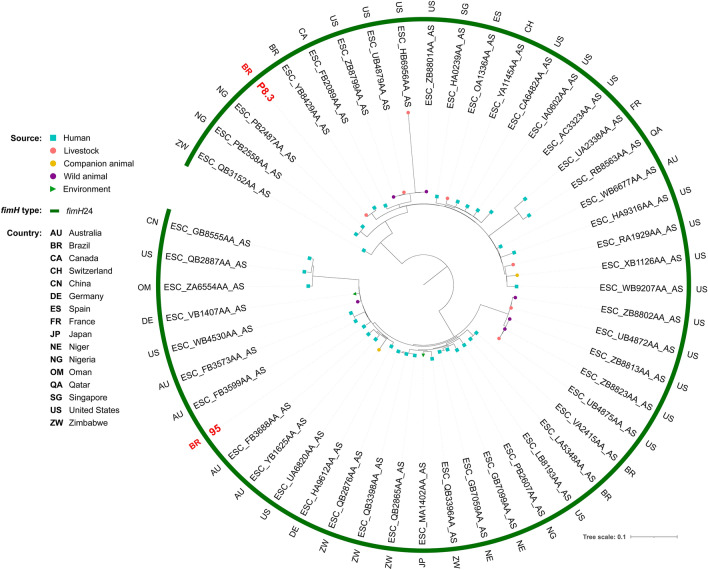
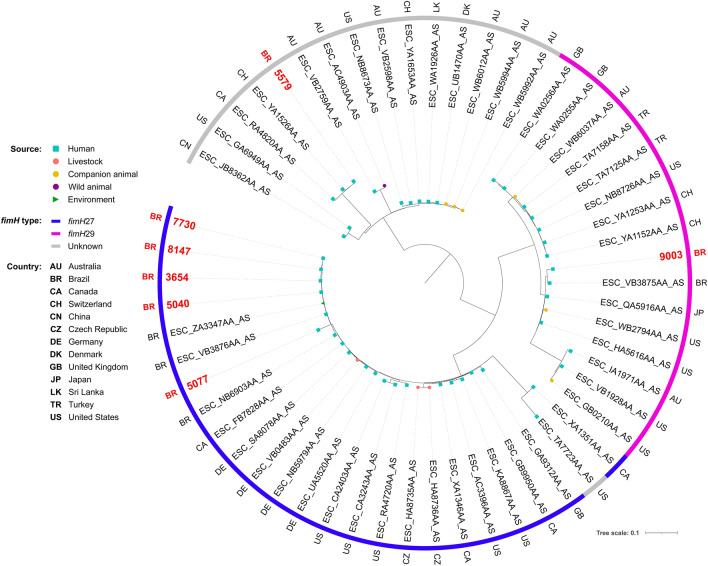
Phylogenetic analysis revealed genomic diversity among isolates. In fact, most of our isolates of the same ST grouped within different clades, closer to isolates from several different countries. For ST38, isolates grouped within two different clades, both mainly composed by strains of human sources (Fig. 2). While two chicken isolates grouped together within a clade with only strains of poultry sources, the other grouped within a clade mainly composed by strains from livestock and wild animal origin. For ST117, most of the isolates were from poultry origin, including closely related strains from human host (Fig. 3). For ST131, while the strain from chicken grouped with strains from various animal sources, human isolates grouped mainly with other human isolates, but isolates were relatively distant to each other, being closer to strains from several other countries (Fig. 4). For the ST354, three of human isolates grouped together within a clade shared only with human isolates, whereas other human isolates clustered into a clade, mainly composed by poultry strains. The isolate from food grouped within a clade with human, poultry, environment and food isolates (Fig. 5). For ST410, the human isolate grouped within a clade shared mainly with strains from human sources, and the isolate from swine grouped within a more diverse clade (Fig. 6). For ST648, human isolates grouped within three different clades, all of them composed mainly by isolates from human origin (Fig. 7).
Figure 2.
Phylogenetic tree of 54 Escherichia coli ST38 strains, their source, country of collection and fimH type.
Figure 3.
Phylogenetic tree of 46 Escherichia coli ST117 strains, their source, country of collection and fimH type.
Figure 4.
(A) Unscaled phylogenetic tree with the source and fimH type of 88 Escherichia coli ST131strains. (B) Subtree containing fimH30 and fimH487 isolates showed in (A), their source, fimH type and country of collection. (C) Subtree containing fimH22 isolates showed in (A), their source, fimH type and country of collection.
Figure 5.
Phylogenetic tree of 43 Escherichia coli ST354 strains, their source, country of collection and fimH type.
Figure 6.
Phylogenetic tree of 51 Escherichia coli ST410 strains, their source, country of collection and fimH type.
Figure 7.
Phylogenetic tree of 56 Escherichia coli ST648 strains, their source, country of collection and fimH type.
Discussion
This study presents the first reports of E. coli ST131-C1-M27 in human infection and CTX-M-24-positive E. coli ST354 from UTI, in Brazil. In this country CTX-M-producing E. coli are endemic. Our data show a wide distribution of these isolates belonging to international clones in livestock and community patients. The extensive presence of CTX-M enzyme-producing strains in several sources raises the hypothesis that the spread occurs with greater frequency and efficiency, especially among Enterobacterales10.
E. coli ST131 is globally disseminated and related to the spread of resistance genes, including specific CTX-M variants17. Recent studies have shown that ST131 is rare among animal isolates, becoming almost exclusively a human pathogen, as demonstrated by our results, where ST131 was predominantly found a human lineage isolated from urine18. The blaCTX-M-15 gene is frequently found in E. coli ST131 UTI and is strongly related to the C2 subclade11. Different groups of plasmids can carry the blaCTX-M-15 gene, here we observe that this gene is involved with the incompatibility group IncF. In a previous study was shown that clade C was related to highest rates of UTI, with subclade C2 being the most common and associated with incompatibility group IncFII19. Besides, CTX-M-15-producing E. coli ST131 has already been shown to be involved in outbreaks in health institutions and is the most prevalent ESBL-producing E. coli worldwide20.
The CTX-M-27-producing ST131-C1 has been considered a new epidemic clone, and it has not been reported in human infections so far, in Brazil. Clade C1-M27 is associated with CTX-M-27 and was first observed colonizing children in France in 2012. Birgy et al. (2019)21 suggests that the C1-27 subclade was recently emerged, because it was observed that the difference in SNPs between the C1 subclade isolates was smaller compared to the SNPs difference between the C2 subclade and the A clade isolates. In addition, the plasmid predominantly involved with the dissemination of blaCTX-M-27 is IncF[F1:A2:B20], as found in our study. Resistance to fluoroquinolones, macrolides, tetracyclines, aminoglycosides, and sulfonamides appears to be part of the profile of C1-M27 isolates21,22.
CTX-M-14 and CTX-M-24 ESBL variants belong to the CTX-M-9 group. Although the first one is widely distributed, especially in China, South-East Asia, Japan, South Korea, and Spain, microorganisms producing CTX-M-24 remain relatively rare, being reported with greater incidence in countries such as Peru and Bolivia19,23,24. This study found an association between CTX-M-24 and E. coli ST354 detected in two human isolates, never before reported in UTI in Brazil. Strains of ST354 isolates were positive to blaCTX-M-24 and displayed further resistance to ciprofloxacin, being associated with extra-intestinal infections, animals and humans, reinforcing the zooanthroponotic hypothesis of these clones25.
CTX-M-2 and CTX-M-55 ESBL variants are commonly reported26. The co-production of CTX-M-8 and CTX-M-55 is not a common phenomena, which was first described in poultry samples in Brazil, in 201812. In this regard the blaCTX-M-8 gene has been located in plasmids IncI1, and the blaCTX-M-55 gene in plasmids IncF12, being confirmed by our results. Therefore, the coexistence of CTX-M-8 and CTX-M-55 in two of our human urine isolates belonging to ST1725 can be explained by the presence of plasmids IncF[F33:A-:B1] and IncI1[ST-113], whereas in the chicken meat isolate belonging to ST6448 we detected IncF[F24:A-:B73] and IncI1[ST-131]23. In the last ten years, IncI1-type plasmids have had a high spread, mainly in animal reservoirs. There are reports of blaCTX-M-2, blaCTX-M-8 and blaCTX-M-55 genes frequently found on IncI plasmids from E. coli isolated from chickens and pigs several countries, such as China, France, the United States of America, and the United Kingdom27–29.
The international clone ST117 was the only ST found in all our three sources studied. This clone is often associated with AMR and found in animals and human isolates, having a zoonotic profile30–32. E. coli ST117 isolates have been identified in pigs, with the blaCTX-M-55 gene being carried by IncN-types plasmids. These data found in our study raises concern, as they may indicate a possible new epidemic transmission of CTX-M-55 in Brazil31,33.
ST38 was found in chicken meat and urine samples. Considered a minority among ESBL-producing E. coli in Europe, Middle East and Asia, the ST38 E. coli has been previously associated with blaCTX-M-2, blaCTX-M-9 and blaCTX-M-14 genes, but rarely with the blaCTX-M-27 gene34,35. The blaCTX-M-27 gene has been associated with IncF plasmids, with the most commonly identified being IncF[F-:A-:B33]35. Interestingly, in our study a ST38 E. coli isolate from human urine carried the blaCTX-M-27 and had plasmid IncF[F2:A-:B10]. Furthermore, recent reports linked CTX-M-types producing E. coli ST38 to UTI36. These data may indicate an increased frequency with which ST38 isolates are being found, being a prominent ESBL lineage worldwide34–36.
In summary, E. coli carrying blaCTX-M genes from different sources seem to be related to the spread of internationally known clones (ST38, ST117, ST131, ST354, ST410, ST648). Some clones associated with CTX-M variants are more prevalent in some sources than others, which does not exclude the possibility that new clones are entering and establishing themselves in different niches. In fact, we detected E. coli ST131-C1-M27 in human infections, and CTX-M-24 positive E. coli ST354 from UTI, which could be a public health issue. Furthermore, the presence of CTX-M (-55, -27, -24, -15, -14 and -2)-producing E. coli among ST38, ST117, ST131 and ST354 clones from humans and meat for consumption reinforces the need for additional surveillance studies, in order to study dynamics of dissemination of ESBL-producing E. coli clones at the human-animal-food-environmental interface.
Material and methods
E. coli isolated from human urine samples
During June 2016 to May 2019, 195,080 urine cultures were performed in a public health services, in a city in south of Brazil. A total of 34,293 (17.6%) were positive for Gram-positive or Gram-negative microorganisms; of these 22,698 (66.2%) were E. coli strains and a total of 2033 (6.2%) ESBL producing bacteria, being 1389 (51.2%) ESBL-producing-E. coli.
As screening criteria for performing the ERIC-PCR, we selected 1,389 E. coli isolates from human urine that presented ESBL production and/or were resistant to quinolones, previously identified by the automated VITEKⓇ 2 system.
Through the analysis of the dendrogram constructed by the ERIC-PCR technique, we selected 59 E. coli isolates from human urine that presented an interesting genetic similarity profile to perform the complete genome sequencing.
All participants provided the written informed consent for this study and the study was approved by the Ethics and Research Committee of the State University of Londrina by a document numbered as CAAE 56869816.0.0000.5231. Besides, we confirm that all experiments were performed in accordance with ethical regulations.
E. coli isolated from chicken meat and pork samples
A surveillance study from January to May 2019 was carried out, to research ESBL-producing E. coli, in chicken and pork meat, bought at markets and butcher shop near public health services. Quinolone-resistant and ESBL-producing E. coli were investigated in chicken meat (n = 50), and pork (n = 50) samples. A total of 102 E. coli was isolated from chicken meat marketed, with 52 ESBL positive. And 67 resistant E. coli were isolated in pork meat, 31 ESBL positive.
We performed ERIC-PCR on all 169 E. coli isolates derived from chicken meat and pork that showed ESBL production and/or quinolone resistance. Through dendrogram analysis, we selected 32 E. coli isolates from chicken meat (n = 24) and pork (n = 8) that presented an interesting genetic similarity profile to perform the complete genome sequencing.
Study population
The study relies on the complete sequencing of 91 E. coli isolates derived from human urine (n = 59), chicken meat (n = 24) and pork (n = 8).
Microbiological methods
All the 195,080 urines collected from women patients during June 2016 to May 2019 was inoculated on CHROMagar (Becton Dickinson, Heidelberg, Germany) and MacConkey (Merck, Darmstadt, Germany) plates using a calibrated inoculating loop with a capacity of 10 μl and incubated at 37 °C for 24 h.
The samples of chicken meat (n = 50) and pork (n = 50) were dipped in Brain Heart Infusion (BHI) broth (Oxoid) with cefotaxime (4 µg/mL), ciprofloxacin (4 µg/mL), and both (Sigma-Aldrich, Munich, Germany) to selected resistant E. coli strains. After incubation, the solution was inoculated in the same way used for urine samples. We evaluated the growth of E. coli from the three BHI solutions and we preferentially selected the isolates that grew in broth with both antimicrobials (cefotaxime and ciprofloxacin). In case there was no growth of any isolate on that broth, we selected from the broth containing separate cefotaxime and ciprofloxacin. All the isolates selected were stored in Tryptic Soy Broth (TSB) with 15% glycerol (− 20◦C).
The identification and bacterial susceptibility were performed by the automated VITEKⓇ 2 system, using the VITEKⓇ 2 AST 239 card and the VITEKⓇ 2 GN ID card (BioMérieux, USA). The bacterial susceptibility was tested for 14 antibiotics: ampicillin, amoxicillin/clavulanate, ceftriaxone, cefepime, ertapenem, meropenem, nalidixic acid, ciprofloxacin, norfloxacin, gentamicin, amikacin, nitrofurantoin, trimethoprim-sulfamethoxazole, and piperacillin-tazobactam. The CLSI 2020 (Clinical and Laboratory Standards Institute) criteria were used for interpretation. E. coli ATCCⓇ25922 strain was used as quality control.
All the time of study, samples were phenotypically confirmed for ESBL production using the chromogenic agar medium (ChromID ESBL bioMèrieux, Marcy L’Etoile, France).
ERIC-PCR
1389 ESBL-producing isolates were subjected to Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR), by Versalovicet al. (1991)37. Analysis of genomic fingerprinting was performed using GelJ v.2.0 software by the Dice similarity method (HERAS et al., 2015)38. Strains were considered genetically related if the similarity index was ≥ 85%.
DNA isolation and whole-genome sequencing
For DNA extraction, strains were grown on Mueller–Hinton Agar overnight at 37 °C. Subsequently, a single colony was inoculated in 2 mL of Luria–Bertani broth for 12 h at 37 °C. The suspension was used to continue extraction and purification by the DNA extraction kit (Invitrogen, Carlsbad, CA). The extracted DNA was quantified by Qubit dsDNA (double-stranded DNA) BR assay kit (Invitrogen, Carlsbad, CA). After quantification, the DNA was used to construct a paired-end library (150 bp), sequenced using the NextSeq platform (Illumina). The instructions of each manufacturer were followed in all steps.
Bioinformatic analysis
Genome quality filter and assemblies were performed by the CLC Genomics Workbench version 7.0 (Aarhus, Denmark). Multilocus sequence type (MLST), resistome, and virulome were identified using MLST v2.0 (Larsen et al., 2012), ResFinder v3.1(Bortolaia et al., 2020), VirulenceFinder v2.0, (Joensen et al., 2014), PlasmidFinder v2.1 (Carattoli et al., 2014), FimTyper v1.0 (Roer et al., 2017) and SerotypeFinder v.2.0 (Joensen et al., 2015), respectively. The BacMet database (Pal et al., 2013) was used to identify biocides and heavy metal (HM)32,39–45.
In order to compare our strains belonging to ST38, ST117, ST131, ST354, ST410 and ST648 with other genetically related strains from different sources and countries, we performed a search for each ST on the Enterobase Escherichia/Shigella database46 (https://enterobase.warwick.ac.uk), then we downloaded all genome assemblies with data for country, source and collection year. Selection of assemblies to phylogenetic analysis was based on fimH type, obtained from Enterobase, and on average nucleotide identity (ANI), assessed with FastANI v1.3247 (https://github.com/ParBLiSS/FastANI).
CSI Phylogeny v1.448 (https://cge.cbs.dtu.dk/services/CSIPhylogeny) was used with default settings to generate phylogenetic trees for each ST, using as reference genome the chromosome sequences of E. coli ST38 strain 144 (RefSeq accession numberNZ_CP023364.1), ST117 strain 14EC020 (NZ_CP024138.1), ST131 strain p4A (NZ_CP049085.2), ST354 strain SMS-3-5 (NC_010498.1), ST410 strain Ecol_517 (NZ_CP018965.1) and ST648 strain Ecol_881 (NZ_CP019029.1).
Phylogenetic trees were rooted at midpoint and annotated with data from Enterobase using iTOL v6 (https://itol.embl.de). Due to the great diversity of ST131, the tree was split into two subtrees, one for fimH22 and the other for fimH30 and fimH485.
Ethical approval
The study was approved by the Ethics and Research Committee of the State University of Londrina CAAE 56869816.0.000.5231.
Supplementary Information
Acknowledgements
This work was completely supported by the Bill & Melinda Gates Foundation (number OPP1193112). Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission. The Grant is part of Grand Challenges Explorations Brazil—New Approaches to Characterize the Global Burden of Antimicrobial Resistance (OPP1193112). Acknowledgments to Araucaria Foundation, the Research Support Facilities Center of the State University of São Paulo (CEFAP-USP), Institute of Biomedical Sciences of State University of São Paulo (ICB-USP) and Master’s program Clinical and Laboratory Pathophysiology of the State University of Londrina.
Author contributions
J.G.M.S., V.L.K., A.T.T., Z.N.T., G.N., R.K.T.K. and E.C.V.: responsible for the collection of samples and their processing. J.G.M.S., L.C. and E.S.: responsible for bioinformatics analyses. J.G.M.S., C.A.M.A., N.L. and E.C.V.: responsible for the construction of the text. All authors were responsible for reviewing the text.
Data availability
Draft whole-genome assemblies were deposited in GenBank under the BioProjectPRJNA578368. The data of the figures can be accessed in Figshare (https://doi.org/10.6084/m9.figshare.12808439.v1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-13197-y.
References
- 1.Poirel L, et al. Antimicrobial resistance in Escherichia coli in antimicrobial resistance in bacteria from livestock and companion animals. Am. Soc. Microbiol. 2018;6(4):289–316. [Google Scholar]
- 2.Reid CJ, McKinnon J, Djordjevic SP. Clonal ST131-H22 Escherichia coli strains from a healthy pig and a human urinary tract infection carry highly similar resistance and virulence plasmids. Microb. Genomics. 2019;5:0013. doi: 10.1099/mgen.0.000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clifford K, et al. Antimicrobial resistance in livestock and poor quality veterinary medicines. Bull. World Health Organ. 2018;96:662–664. doi: 10.2471/BLT.18.209585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gozi KS, et al. Dissemination of multidrug-resistant commensal Escherichia coli in feedlot lambs in southeastern Brazil. Front. Microbiol. 2019;10:1394. doi: 10.3389/fmicb.2019.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paphitou NI. Antimicrobial resistance: Action to combat the rising microbial challenges. Int. J. Antimicrob. Agents. 2013;42:S25–S28. doi: 10.1016/j.ijantimicag.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Cyoia PS, et al. Distribution of ExPEC virulence factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 2019;10:1–9. doi: 10.3389/fmicb.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daga AP, et al. Escherichia coli bloodstream infections in patients at a university hospital: virulence factors and clinical characteristics. Front. Cell. Infect. Microbiol. 2019;9:191. doi: 10.3389/fcimb.2019.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irrgang A, et al. CTX-M-15-producing E. coli isolates from food products in Germany are mainly associated with an IncF-type plasmid and belong to two predominant clonal E. coli lineages. Front. Microbiol. 2017;8:2318. doi: 10.3389/fmicb.2017.02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonçalves LF, et al. Multidrug resistance dissemination by extended-spectrum β-lactamase-producing Escherichia coli causing community-acquired urinary tract infection in the Central-Western Region, Brazil. J. Glob. Antimicrob. Resist. 2016;6:1–4. doi: 10.1016/j.jgar.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Silva N, et al. Genetic characterisation of extended-spectrum β-lactamases in Escherichia coli isolated from retail chicken products including CTX-M-9 containing isolates: A food safety risk factor. Br. Poult. Sci. 2012;53:747–755. doi: 10.1080/00071668.2012.740554. [DOI] [PubMed] [Google Scholar]
- 11.Coque TM, et al. Dissemination of clonally related E. coli strains expressing ESBL CTX-M-15. Emerg. Infect. Dis. 2008;14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes MR, Sellera FP, Moura Q, Souza TA, Lincopan N. Draft genome sequence of a CTX-M-8, CTX-M-55 and FosA3 co-producing Escherichia coli ST117/B2 isolated from an asymptomatic carrier. J. Glob. Antimicrob. Resist. 2018;12:183–184. doi: 10.1016/j.jgar.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Peirano G, et al. Trends in population dynamics of Escherichia coli sequence type 131, Calgary, Alberta, Canada, 2006–20161. Emerg. Infect. Dis. 2020;26:2907–2915. doi: 10.3201/eid2612.201221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flament-Simon SC, et al. High prevalence of ST131 subclades C2–H30Rx and C1–M27 among extended-spectrum β-lactamase-producing Escherichia coli causing human extraintestinal infections in patients from two hospitals of Spain and France during 2015. Front. Cell. Infect. Microbiol. 2020;10:1–9. doi: 10.3389/fcimb.2020.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumura Y, et al. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg. Infect. Dis. 2016;22:1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duggett N, et al. Detection in livestock of the human pandemic Escherichia coli ST131 fimH30(R) clone carrying blaCTX-M-27. J. Antimicrob. Chemother. 2021;76:265–267. doi: 10.1093/jac/dkaa407. [DOI] [PubMed] [Google Scholar]
- 17.Kondratyeva K, Salmon-Divon M, Navon-Venezia S. Meta-analysis of pandemic Escherichia coli ST131 plasmidome proves restricted plasmid-clade associations. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-019-56763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pitout JDD, DeVinney R. Escherichia coli ST131: A multidrug-resistant clone primed for global domination. F1000Research. 2017;6:1–7. doi: 10.12688/f1000research.10609.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peirano G, Pitout JDD. Extended-spectrum β-lactamase-producing Enterobacteriaceae: Update on molecular epidemiology and treatment options. Drugs. 2019;79:1529–1541. doi: 10.1007/s40265-019-01180-3. [DOI] [PubMed] [Google Scholar]
- 20.Nicolas-Chanoine MH, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008;61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 21.Birgy A, et al. Independent host factors and bacterial genetic determinants of the emergence and dominance of Escherichia coli sequence type 131 CTX-M-27 in a community pediatric cohort study. Antimicrob. Agents Chemother. 2019;63:1–10. doi: 10.1128/AAC.00382-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandes MR, et al. Emergence of CTX-M-27-producing Escherichia coli of ST131 and clade C1–M27 in an impacted ecosystem with international maritime traffic in South America. J. Antimicrob. Chemother. 2020;75:1647–1649. doi: 10.1093/jac/dkaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J, Oh SS, Kim J, Shin J. Extended-spectrum β-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea. Sci. Rep. 2020;10:1–7. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delgado DYC, Barrigas ZPT, Astutillo SGO, Jaramillo APA, Ausili A. Detection and molecular characterization of β-lactamase genes in clinical isolates of Gram-negative bacteria in Southern Ecuador. Braz. J. Infect. Dis. 2016;20:627–630. doi: 10.1016/j.bjid.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dagher C, et al. Molecular characterization of carbapenem resistant Escherichia coli recovered from a tertiary hospital in Lebanon. PLoS ONE. 2018;13:1–13. doi: 10.1371/journal.pone.0203323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunha MPV, et al. Coexistence of CTX-M-2, CTX-M-55, CMY-2, FosA3, and QnrB19 in extraintestinal pathogenic Escherichia coli from poultry in Brazil. Antimicrob. Agents Chemother. 2017;61:1–3. doi: 10.1128/AAC.02474-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J, Oh SS, Kim J, Park S, Shin J. Clinically relevant extended-spectrum β-lactamase–Producing Escherichia coli isolates from food animals in South Korea. Front. Microbiol. 2020;11:1–9. doi: 10.3389/fmicb.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castellanos LR, et al. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian poultry chain. PLoS ONE. 2017;12:1–15. doi: 10.1371/journal.pone.0170777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri Junior A, Darini ALC. Evaluation and characterization of plasmids carrying CTX-M genes in a non-clonal population of multidrug-resistant Enterobacteriaceae isolated from poultry in Brazil. Diagn. Microbiol. Infect. Dis. 2016;85:444–448. doi: 10.1016/j.diagmicrobio.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Cormier A, et al. Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet. Microbiol. 2019;231:71–75. doi: 10.1016/j.vetmic.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 31.Norizuki C, et al. Detection of Escherichia coli producing CTX-M-1-group extended-spectrum β -lactamases from pigs in Aichi Prefecture, Japan, between 2015 and 2016. Jpn. J. Infect. Dis. 2018;71:33–38. doi: 10.7883/yoken.JJID.2017.206. [DOI] [PubMed] [Google Scholar]
- 32.Lupo A, Saras E, Madec JY, Haenni M. Emergence of bla CTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 2018;73:867–872. doi: 10.1093/jac/dkx489. [DOI] [PubMed] [Google Scholar]
- 33.Xia L, et al. Prevalence of ST1193 clone and IncI1/ST16 plasmid in E. coli isolates carrying blaCTX-M-55 gene from urinary tract infections patients in China. Sci. Rep. 2017;7:1–8. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi W, et al. High prevalence of blaCTX-M-14 among genetically diverse Escherichia coli recovered from retail raw chicken meat portions in Japan. Int. J. Food Microbiol. 2018;284:98–104. doi: 10.1016/j.ijfoodmicro.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Mostafa, H. H. et al. Genomic surveillance of ceftriaxone-resistant Escherichiacoli in western new uork suggests the extended-spectrum β-lactamase blaCTX-M-27 is emerging on distinct plasmids in ST38. Front. Microbiol.11, 1747 (2020). [DOI] [PMC free article] [PubMed]
- 36.Birgy A, et al. Diversity and trends in population structure of ESBL-producing Enterobacteriaceae in febrile urinary tract infections in children in France from 2014 to 2017. J. Antimicrob. Chemother. 2020;75:96–105. doi: 10.1093/jac/dkz423. [DOI] [PubMed] [Google Scholar]
- 37.Versalovic J, Koeuth T, Lupski JR, Plaza OB. Institute for Molecular Genetics and Department of Pediatrics, Baylor College of Medicine. Cell. 1991;19:6823–6831. [Google Scholar]
- 38.Heras J, et al. GelJ—A tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015;16:1–8. doi: 10.1186/s12859-015-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen MV, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bortolaia V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinheinz KA, Joensen KG, Larsen MV. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carattoli A, et al. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roer L, et al. Development of a web tool for Escherichia coli subtyping based on fimh alleles. J. Clin. Microbiol. 2017;55:2538–2543. doi: 10.1128/JCM.00737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DGJ. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014;42:737–743. doi: 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:1–8. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:1–8. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Draft whole-genome assemblies were deposited in GenBank under the BioProjectPRJNA578368. The data of the figures can be accessed in Figshare (https://doi.org/10.6084/m9.figshare.12808439.v1).