Abstract
BACKGROUND AND AIMS:
Hepatocellular carcinoma (HCC) is an aggressive malignancy which is often associated with a complex tumor microenvironment attributable to etiology-induced cellular inflammation. γδ T cells are known to detect and react to chronic inflammation, which is linked to cancer development, progression, and metastasis. Our recent genomic study revealed an increased infiltration of several immune cell types, including γδ T cells, in tumor microenvironments of a Thai HCC subtype associated with a good prognosis.
APPROACH AND RESULTS:
Here, we quantified the amount of γδ T cells using a γδ T-cell-specific gene signature in 247 Chinese HCC patients. We also validated the γδ T-cell signature in American HCC patients. Additionally, such an association was only found in tumor transcriptomic data, but not in adjacent nontumor transcriptomic data, suggesting a selective enrichment of γδ T cells in the tumor microenvironment. Moreover, the γδ T-cell signature was positively correlated with the expression of natural killer cell receptor genes, such as NKG2D and cytolytic T-cell genes granzymes and perforin, suggesting a stronger T-cell-mediated cytotoxic activity. Furthermore, we found that the γδ T-cell-specific gene expression is positively correlated with the expression of chemokine (C-C motif) ligand 4 (CCL4)/chemokine (C-C motif) ligand 5 (CCL5) and C-C chemokine receptor type 1 (CCR1)/C-C chemokine receptor type 5 (CCR5), the receptors for γδ T cells. We validated these results using immunohistochemical analysis of formalin-fixed, paraffin-embedded tumor biopsies from 182 HCC patients. Moreover, we found evidence of CCL4/CCL5-mediated recruitment of γδ T cells both in vitro and in a murine orthotopic Hepa1–6 HCC model.
CONClUSIONS:
We propose that CCL4/CCL5 may interact with their receptor, CCR1/CCR5, which may facilitate the recruitment of γδ T cells from peripheral blood or peritumor regions to the tumor regions. Consequently, an increasing infiltration of γδ T cells in tumors may enhance antitumor immunity and improve patients’ prognosis.
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide and accounts for 90% of primary liver cancer.(1) HCC portends a poor prognosis given the frequency of late-stage diagnosis and a high rate of tumor recurrence following curative resection.(2) Chronic inflammation plays a major role in the development of HCC and response to therapy.(3) Moreover, accumulating evidence indicates that immune-cell dysregulation occurs in HCC patients, which includes an exacerbated inflammatory environment. Dysregulation is evident by an increased infiltration of leukocytes, such as macrophages and T cells, in addition to our recent findings on γδ T cells and a skewed cytokine and chemokine profile.(4–7) γδ T cells belong to a specialized T-cell subset known to play an important role in immune homeostasis by detecting and reacting to chronic inflammation.(8) However, its role in cancer development and tumor metastasis, especially HCC, is largely unknown. γδ T cells, which consist of γ- and δ-c hain T-cell receptor (TCR) heterodimers, represent 2%−10% of all human T lymphocytes in the peripheral blood.(9) γδ T cells are found in a variety of tissues, most abundantly in the intestinal epithelial lining.(10) Intestinal venous return through the portal system then allows for γδ T cells to travel to the liver, where they comprise up to 35% of all T cells. Although the liver is considered an immunological organ with predominant innate immunity, the role that γδ T cells play has yet to be fully elucidated.
Previous studies have revealed that γδ T cells have high levels of the non-major histocompatibility complex–r estricted cytotoxicity activity against primary hepatocytes and produce high levels of interleukin-8, interferon (IFN)-γ, and tumor necrosis factor alpha in liver biopsies from hepatitis C virus (HCV)- or hepatitis B virus (HBV)-patients.positive patients compared to nonvirally infected (10,11) Moreover, γδ T cells could play some immune regulation effects given that increased numbers of γδ T cells have been identified in the circulation of patients with stable liver graft function, and the total circulating peripheral γδ T-cell populations in patients are increased after liver transplantation.(12–14) Thus, although not a dominant T-cell infiltrate in the liver, the γδ T-cell population has been found to be enriched in livers of patients with liver disease. Recently, by performing systems integration of genomics, transcriptomics, and metabolomics, we have identified an Asian-specific molecular subtype of HCC, namely HCC-C2, which can be found in Thai, Chinese, and Asian American patients, but not in patients of European decent.(7) This subtype is associated with an increased body mass index, elevated bile acid metabolism, and leukocyte infiltration that includes γδ T cells and CD4+ memory T cells.(7) Increased infiltrating T cells in the HCC-C2 subtype also suggest a plausible scenario whereby this tumor type may be sensitive to immune checkpoint inhibitors. To further explore the role of γδ T cells in hepatocarcinogenesis, we determined the correlation between γδ T-cell activity and HCC prognosis.
Materials and Methods
CLINICAL SPECIMENS
A described cohort of 247 Chinese HCC patients, obtained with informed consent from the Liver Cancer Institute (LCI) and Zhongshan Hospital (Fudan University, Shanghai, China) with publicly available Affymetrix U133A array data (National Center for Biotechnology Information Gene Expression Omnibus accession number GSE14520), was used to evaluate the prognostic correlation of γδ T-cell proportions for HCC patients.(15) The study was approved by the Institutional Review Board of the participating institutes. For 240 patients, disease-free and overall survivals (OSs) as well as the cause of death were available.(15) Among them, a subset of patients had paraffin-embedded, formalin fixed (FFPE) blocks used for tissue microarray (TMA) described below. In the validation analysis, The Cancer Genome Atlas (TCGA) Liver Cancer transcriptome data were used.(16) There were 366 total patients, but only 242 patients with survival data were included.
TISSUE IMMUNOHISTOCHEMICAL STAINING AND EVALUATION
TMA was constructed as described(17) and a total of 182 tumor and 114 nontumor blocks were included. Briefly, core samples were punched from representative regions of each donor block according to hematoxylin and eosin (H&E)-stained slides. Tissue cylinders of 1.0 mm in diameter were arrayed on a recipient paraffin block using a tissue arrayer (Pathology Devices, Westminster, MD). For the assessment of TCRγδ and regulated on activation, normal T expressed and secreted (RANTES; chemokine (C-C motif ) ligand 5 [CCL5]) expression, 5-μ m TMA sections were used for immunohistochemical (IHC) staining. TMA sections were deparaffinized with xylene and graded alcohols and antigen recovery was performed in heat-activated antigen retrieval buffer of pH 9.0 (for TCR) or pH 6.0 (for CCL5; Dako, Carpinteria, CA). Standard two-step protocol was applied with primary antibodies: mouse antihuman TCRγδ (Clone H-41, diluted 1:150; Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-RANTES (CCL5; Clone A-4, diluted 1:200; Santa Cruz Biotechnology) incubated at 4°C overnight. Sections were serially rinsed and incubated with secondary antibody according to the manufacturer’s recommendation (Dako). Tissue sections were lightly counterstained with hematoxylin, dehydrated in ethanol, and cleared in xylene. Finally, slides were coverslipped and scanned by the NanoZoomer 2.0 HT (Hama-matsu Photonics K.K., Japan). The number of positive staining of TCRγδ cells in each sample were counted by two pathologists independently. The IHC score of CCL5 was calculated by multiplying the percentage of cells positively stained (0, 1, 5, 10, 20, 30, 40, 50, 60, 70, 80, 90, or 100%) by the intensity of staining (0 = negative, 1 = weak, 2 = intermediate, and 3 = strong) with a maximum of 300.(18)
γδ T CELLS MIGRATION ASSAY
Chemotaxis of γδ T cells was analyzed in transwell migration assays. γδ T cells were expanded and sorted from splenocytes as described.(19) Next, 5 × 105 γδ T cells were taken up in RPMI 1640 and placed into 5-mm pore-size transwell inserts (Corning, NY), which were placed into 24 wells of a tissue-culture plate containing 5 × 105 Hepa1–6 cells with 0.5 μg/mL of anti-CCL5 (R&D MAB478; R&D Systems, Minneapolis, MN) or 3 μg/mL of anti-CCL4 (R&D MAB451). After incubation at 37°C for 120 minutes, γδ T cells migrated to the lower compartment were harvested and quantified by flow cytometry (Canto II; BD biosciences, San Jose, CA), using the Trucount tubes (BD Biosciences, San Diego, CA) and fluorescein-labeled antibody (fluorescein isothiocyanate/anti-CD3, phycoerythrin/cyanine 7/anti-TCR γδ; BioLegend). γδ T cells were identified as CD3+TCR γδ+ cells. The absolute count of γδ T cells/μL = (number of events in quadrant containing cell population/number of events in absolute-count bead region [R2])] × [(total number of absolute-count beads)/(test volume). Migration was evaluated by calculating the percentage of γδ T cells in the bottom well relative to input.
ORTHOTOPIC HCC MOUSE MODEL
Male C57BL/6J mice aged 6–8 weeks were purchased from the Laboratory Animal Center of Chinese Academy of Medical Sciences (Beijing, China). Housing and all procedures were performed according to guidelines from the Institutional Animal Care and Use Committee of Tianjin Medical University. To examine the in vivo immune attractive function of tumor-cell–derived CCL5, we designed CCL5 short hairpin RNA (shRNA) oligos and transferred them into Hepa1–6 cells. Three recommended sequences for CCL5 genes were synthesized, and the most efficient one was used for the relevant assays. Next, 2 × 106 shRNA transfected or nontransfected Hepa1–6 cells were suspended in 60 μL of phosphate-buffered saline containing 50% Matrigel (BD Biosciences). The orthotopic xenograft HCC mice model was created by direct intrahepatic injection of Hepa1–6 cells according to previous reports.(20) Briefly, mice were anesthetized intraperitoneally with 5% chloral hydrate. Fully sedated mice were then placed in a supine position, and a small transverse incision below the sternum was made to expose the liver. Suspended tumor cells were slowly injected into the upper left lobe of the liver using a 28-gauge needle. After injection, light pressure was applied for 1 minute to prevent bleeding. The abdomen was then closed with a 5–0 silk suture. After tumor-cell implantation, animals were kept in a warm cage overnight and returned to the animal room on next morning. At day 14 postinoculation, livers were excised and liver tumor-infiltrated lymphocytes (TILs) were isolated. Flow cyotmetric analysis of the γδ T-cell proportion was used to determine its migration in vivo, and real-time PCR was used to analyze the gene expression level of IFN-γ and perforin.
STATISTICAL ANALYSES
Class comparison and survival risk prediction of the gene expression data were done with the BRB-Array Tools software (Version 4.6.0; Biometric Research Branch, National Cancer Institute, Bethesda, MD). For survival risk prediction, we identified the γδ T-cell-specific gene set from CIBERSORT.(21) High- and low-risk groups of patients were predicted by using Cox’s proportional hazards model based on principle component analysis of the obtained genes with 10-fold cross-validation. The exponential component of Cox’s proportional hazards model was considered as a prognostic index, which can be refined as ∑iwi xi + b, where wi and xi are the weight and logged gene expression of the i-th gene, and b is a constant. A high value of the prognostic index corresponds to a high risk of death, whereas a low value indicates a low risk. Kaplan-Meier curves were provided for the two risk groups. Statistical significance of the association between γδ T-cell-specific genes, and patient survival was further assessed by 1,000 times permutation of survival data, with permutation P value of log-rank test provided.
Kaplan-Meier survival analysis was done using GraphPad Prism software (version 7.0; GraphPad Software Inc., San Diego, CA), and the statistical P values were generated by the Cox-Mantel log-rank test. Cox proportional hazards regression was used to analyze the effect of clinical variables on patient survival, using STATA software (v15; StataCorp LP, College Station, TX). Clinical variables included age, sex, HBV active status, serum alpha-fetoprotein (AFP), cirrhosis, tumor size (the size of the largest tumor when multiple tumors are present), nodular type, microvascular invasion, and the HCC prognosis staging systems, Barcelona Clinic Liver Cancer (BCLC), Cancer Liver Italian Program (CLIP), Okuda staging system (OKUDA), or Tumor Node Metastasis (TNM) classification.(22) An AFP cutoff of 300 ng/mL, carcinoembryonic antigen (CEA) cutoff of 3 ng/mL, cancer antigen 19–9 (CA 19–9) of 37 U/mL, and tumor size of 3 cm were used in the Cox regression analysis and are clinically relevant values used to distinguish patient survival. A univariate test was used to examine the influence of the γδ T-cell gene signature or each clinical variable on patient survival. A multivariate analysis was done to estimate the hazards ratio of the predictor while controlling for clinical variables that were significantly associated with survival in the univariate analysis. Unsupervised hierarchical clustering analysis was performed by the GENESIS software (version 1.8.1) developed by Alexander Sturn (IBMT-TUG, Graz, Austria).
Results
A γδ T-CELL GENE SIGNAT URE PREDICTS SURVIVAL AND RECURRENCE IN HCC
Our recent studies revealed elevated infiltrations of intratumoral γδ T cells linked to Thai patients with an HCC-C2 subtype.(7) To further extend this observation, we first examined the LCI cohort using 55 genes specific for γδ T cells previously defined by CIBERSORT, a method used for characterizing cellular composition for bulk tissues based on gene expression data (http://cibersort.stanford.edu/).(21) The LCI cohort consisted of 240 predominantly HBV- and cirrhosis-positive Chinese HCC patients (Supporting Table S1). To determine a potential clinical significance of the γδ T-cell-related genes, we performed survival risk-prediction analysis using 10-f old cross-validation and tested statistical significance by 1,000-r andom permutation of the survival data. We found that the γδ T-cell-specific genes could stratify HCC patients into two main groups, that is, low risk (LR) and high risk (HR; Fig. 1A), with a significant difference in OS (log-rank, P = 0.0021; Fig. 1B, left panel). The cross-validated misclassification rates were significantly lower than expected by chance (permutation, P = 0.007). However, no significant difference in OS was found in paired nontumor tissues (Fig. 1B, right panel), indicating that the γδ T gene signature is tumor specific. Moreover, this signature was associated with tumor recurrence following resection (P = 0.0034; Fig. 1C). The median recurrence time was 42.2 months in the LR group compared to 14.6 months in the HR group. To further determine the robustness of this signature, we included only the TCGA HCC samples (n = 242) with available OS and recurrence status for further analysis. Consistent with the LCI cohort, the γδ T-cell-specific signature was significantly associated with OS and recurrence in the TCGA HCC cohort. The median survival time for LR was 104.2 months compared to 71.03 months for the HR group, with log-rank P = 0.0036 and permutation P = 0.05. Median recurrence time for LR was 67.60 months compared to 40.33 for HR (P = 0.0033; Supporting Fig. S1A,B). We did not extend our analyses for the nontumor group in the TCGA cohort because of only a small number of samples available.
FIG. 1.
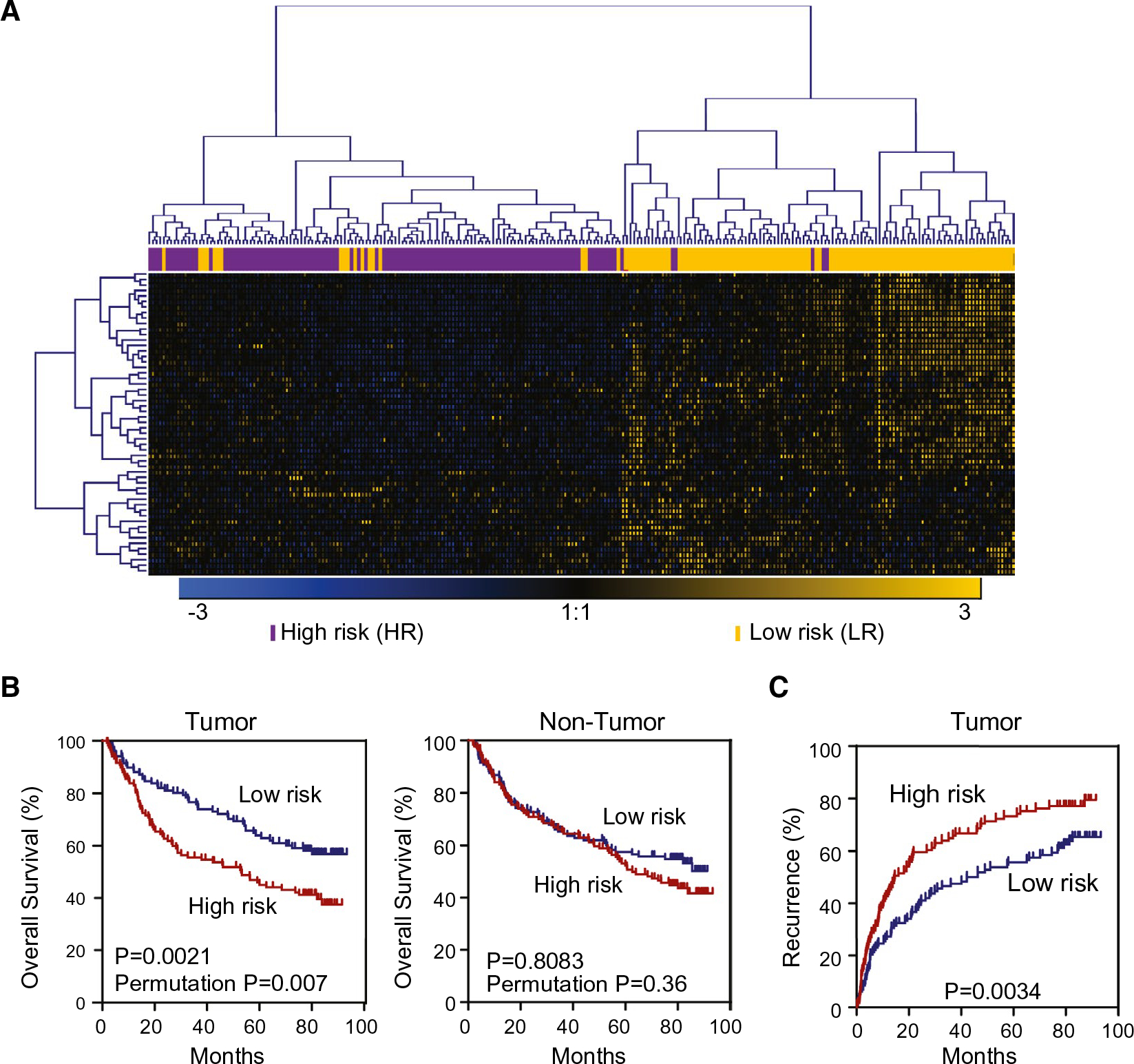
Expression of γδ T-cell-specific genes in tumor tissues, but not nontumor tissues, is associated with HCC prognosis. Based on hierarchical clustering analysis of the 55 γδ T-cell genes signature, patients with HCC were divided into two subgroups: LR and HR. (A) The hierarchical clustering of 55 γδ T-cell-specific genes. Each column represents an individual tissue sample. Genes and samples were ordered by centered correlation and ward linkage. The scale represents gene expression levels from −3.0 to 3.0 in a log2 scale. Each case status is categorized by the γδ T-cell genes signature markers are included above the heatmap. (B) Kaplan-Meier survival analyses of 240 Chinese HCC cases based on survival risk-prediction results of the γδ T-cell gene set in tumor (left panel) and nontumor (right panel). (C) Recurrence-free survival in tumor tissues of 240 Chinese HCC cases described in (B).
We next determined a relative proportion of intratumoral and adjacent infiltrating γδ T cells in both the LCI and TCGA cohorts using CIBERSORT.(21) These analyses revealed that the abundance of γδ T cells was much higher in nontumor tissues than paired tumor tissues (Fig. 2A). Noticeably, a reduction in abundance of γδ T cells in tumor tissues compared to paired nontumor tissues was much greater in the HR group than the LR group (P = 0.0010), whereas there was no significant difference between nontumor tissues of HR and LR groups (P = 0.9914; Fig. 2A). To determine whether γδ T cells were associated with functionally active infiltrating lymphocytes, we measured cytolytic activity based on two key effector genes, that is, granzyme A (GZMA) and perforin 1 (PRF1) in both LCI and TCGA cohorts. These two genes were recently developed by Hacohen et al. as a quantitative measure of the activity of TILs, especially CD8+ T cells where they were significantly elevated upon activation.(23) We found that cytolytic activities were much higher in the LR group than the HR group in both cohorts, indicating that γδ T-cell-associated tumors from the LR group had a much higher immune reactivity than that of the HR group (Fig. 2B and Supporting Fig. S1C). We also compared expression levels of killer cell lectin-like receptor K1 (KLRK1), DNAX accessory molecule 1 (DNAM-1; CD226), and CD96 to determine whether cytolytic activity was associated with increased expression of γδ T-cell-related cytolytic receptors.(24) Gene expression of all three receptors was significantly higher in the LR group than the HR group in both the LCI and TCGA cohorts (P < 0.05; Fig. 2C–E and Supporting Fig. S1D–F). These results indicate that increased intratumoral γδ T-cell infiltration is associated with stronger cytolytic activity of γδ T cells and better antitumor immune status.
FIG. 2.
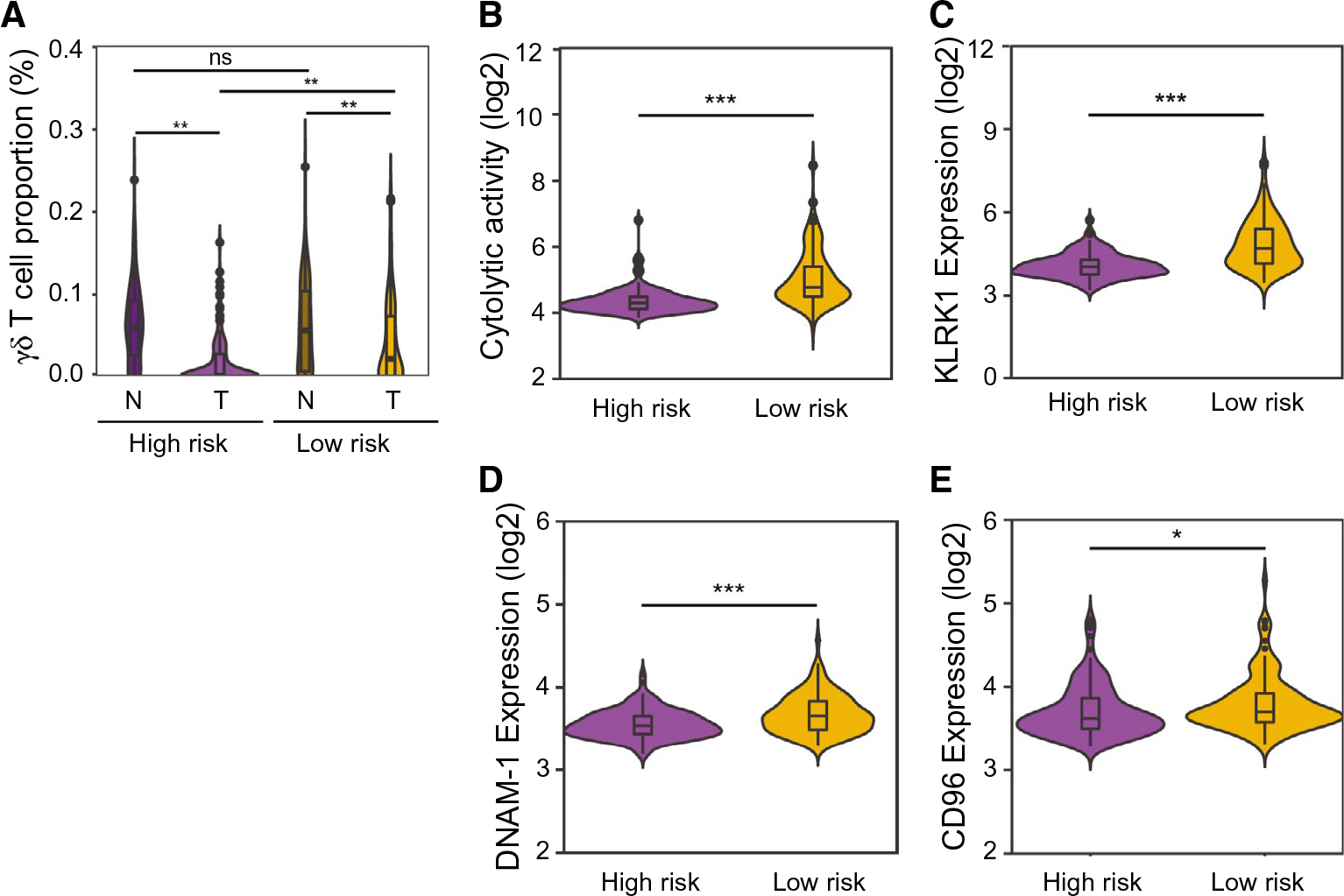
Characteristics of the two subgroups, HR and LR, calculated by the gene transcript data. (A) γδ T-cell proportions in the predefined subgroups of both tumor and nontumor. (B) Immune cytolytic activity (“CYT”) was calculated based on transcript levels of two key cytolytic effectors, GZMA and PRF1. (C) Comparison of KLRK1 expression level in the HR and LR subgroups in HCC tumor samples. (D) Comparison of DNAM-1 (CD226) expression level in the HR and LR subgroups in HCC tumor tissues. (E) Comparison of CD96 expression level in the HR and LR subgroups in HCC tumor samples. *P < 0.05; **P < 0.01; ***P < 0.001.
To further validate our gene array data, we performed IHC analyses using anti-TCRγδ and anti-RANTES (CCL5) in 182 HCC tumor and 114 adjacent nontumor tissue samples of the LCI cohort to determine the distributions of γδ T cells. Positive anti-TCRγδ cells were defined as brown-stained cells scattered in the tumoral and peritumoral parenchyma (Fig. 3A). We found that there were significantly less TCRγδ-positive cells in HCC samples compared to paired nontumor samples (P < 0.001; Fig. 3B). Correlation analysis between transcriptome-based estimates of the presence of γδ T cells and a total number of TCRγδ-positive cells based on IHC analyses revealed a positive correlation (r = 0.3055; P = 0.0197; Fig. 3C), confirming that transcriptome-based approaches are reasonable in quantifying γδ T cells. Consistent with transcriptome data, HCC patients with increased γδ T-cell infiltration had a significantly better prognosis than HCC patients with low γδ T-cell infiltration (Fig. 3D). In contrast, γδ T-cell infiltration in the matched peritumor tissues was not associated with OS (Fig. 3E).
FIG. 3.
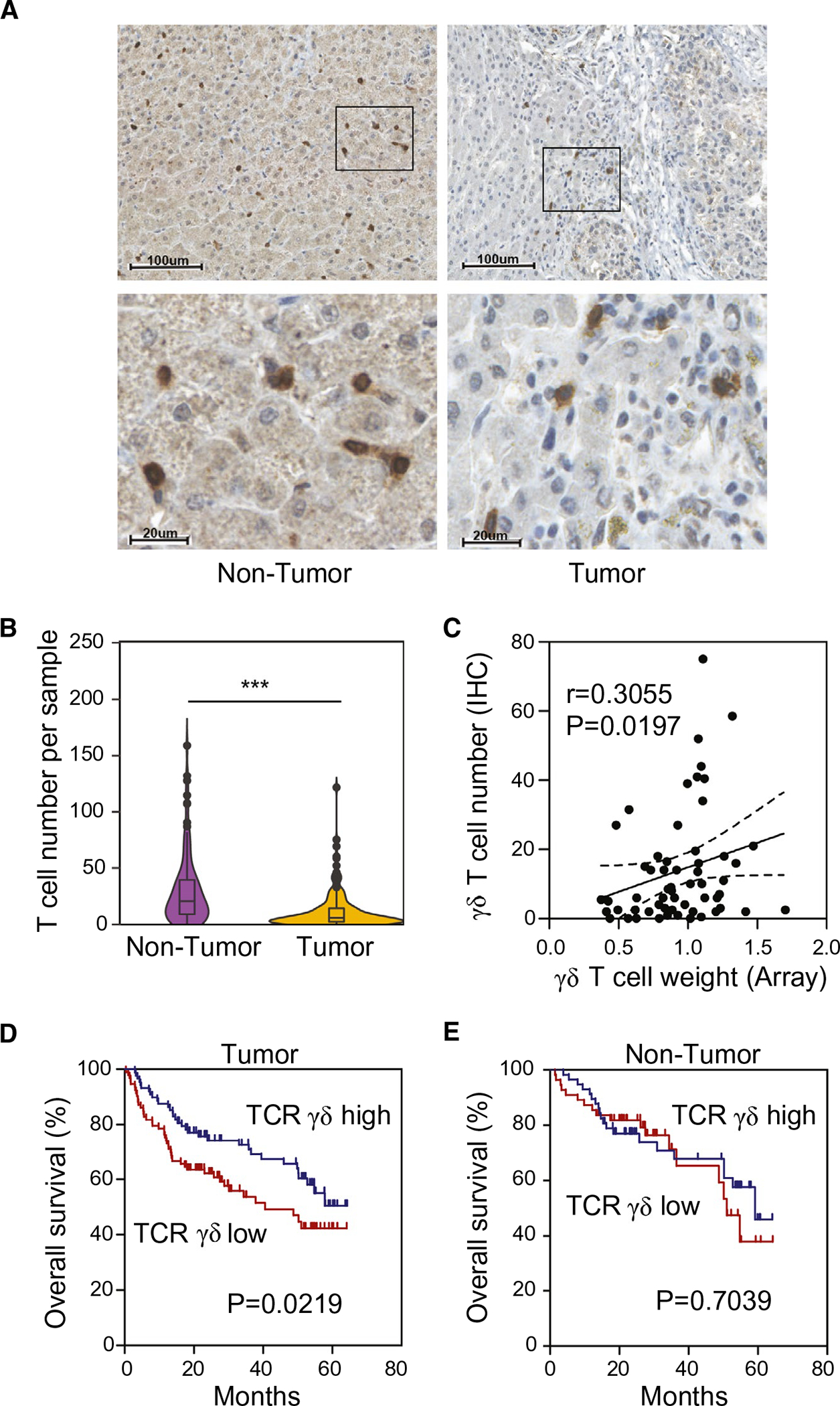
Validation of γδ T-cell expression in the predefined LR and HR subgroups in both HCC tumor and nontumor tissues. Antihuman TCRγδ were used, and the numbers of positive staining of TCRγδ cells in every sample were counted. (A) Representative IHC staining pattern of TCR γδ+ in tumor (right) and paired nontumor (left) tissues. Magnification power in upper panel is 100×, the scale bar represents 100 μm; in lower panel magnification is 400×, and the scale bar represents 20μm. (B) The comparison of the number of TCRγδ-positive staining cells in tumor and paired nontumor tissues detected by IHC detection, ***P < 0.001. (C) Correlation analysis between the γδ T-cell proportion calculated from array data and IHC-positive staining cell counts. (D) Kaplan-Meier survival analyses of 182 HCC tumor samples stratified by TCRγδ expression status, TCRγδ high group (n = 88), and TCRγδ low group (n = 94). (E) Kaplan-Meier survival analyses of 114 nontumor samples stratified by TCRγδ expression status, TCRγδ high group (n = 57), and TCRγδ low group (n = 57).
To determine the clinical characteristics between the LR and HR groups, we performed Cox regression analyses in the LCI cohort. In the LR subgroup, there were less active viral replication chronic carriers and less AFP elevation. In addition, the LR group had lower multinodularity as well as less microvascular invasion (chi-squared test, P < 0.05; Table 1). Univariate Cox analyses revealed that the γδ T-cell gene signature is associated with OS (hazard ratio = 0.57; 95% confidence interval [CI] = 0.40–0.83), as well as other clinical variables, including sex, underlying liver cirrhosis, serum CA 19–9 level, tumor size, microscopic vascular invasion, and tumor staging such as CLIP, BCLC, or TNM (Table 2). Using only significant variables from the univariate analyses, we performed multivariate analyses to determine whether the γδ T-cell gene signature was a predictor of OS independent of other clinical variables. Multivariate Cox regression analyses, adjusting other variables that were significantly associated with OS, revealed that γδ T-cell gene signature remained an independent predictor of OS (Table 2). Together, these results suggest that γδ T cells may play some protective roles in the progression of HCC.
TABLE 1.
Clinical Characteristics of patients in the lCI Cohort according to γδ t Cell Subgroup (n = 240)
| Clinical Variable | LR n = 119 (%) | HR n = 121 (%) | P Value |
|---|---|---|---|
|
| |||
| Sex | 0.886 | ||
| Male | 104 (87.4) | 105 (86.8) | |
| Female | 15 (12.6) | 16 (13.2) | |
| Age, years | 0.999 | ||
| ≤50 | 60 (50.4) | 61 (50.4) | |
| >50 | 59 (49.6) | 60 (49.6) | |
| HBV virus status | 0.036 | ||
| None | 17 (14.2) | 7 (5.8) | |
| Chronic carrier | 83 (69.7) | 75 (62.0) | |
| Active viral replication | 19 (15.9) | 39 (32.2) | |
| AFP | 0.020 | ||
| Normal | 72 (60.5) | 55 (45) | |
| Elevated | 47 (39.5) | 66 (55) | |
| CEA | 0.305 | ||
| Normal | 63 (52.9) | 72 (59.5) | |
| Elevated | 56 (47.1) | 49 (40.5) | |
| CA 19– 9 | 0.714 | ||
| Normal | 74 (62.2) | 78 (64.5) | |
| Elevated | 45 (37.8) | 43 (35.5) | |
| BCLC stage | 0.084 | ||
| 0 | 7 (6.66) | 12 (10.3) | |
| A | 81 (76.4) | 71 (60.7) | |
| B | 7 (6.66) | 16 (13.7) | |
| C | 11 (10.4) | 18 (15.4) | |
| TNM staging | 0.005 | ||
| I | 58 (54.21) | 39 (32.50) | |
| II | 31 (28.97) | 48 (40.00) | |
| III | 18 (16.82) | 33 (27.50) | |
| Okuda staging | 0.414 | ||
| 0 | 92 (87.6) | 99 (83.8) | |
| 1 | 13 (12.4) | 19 (16.2) | |
| CLIP staging | 0.222 | ||
| Early (0–2) | 101 (84.9) | 109 (90.1) | |
| Late (3–5) | 18 (15.1) | 12 (9.9) | |
| Multinodular | 0.047 | ||
| No | 100 (84.0) | 89 (73.5) | |
| Yes | 19 (16.0) | 32 (26.5) | |
| Tumor size, cm | 0.062 | ||
| ≤3 | 43 (36.1) | 30 (25.0) | |
| >3 | 76 (63.9) | 90 (75.0) | |
| Microvascular invasion | 0.021 | ||
| No | 58 (63.0) | 47 (46.5) | |
| Yes | 34 (37.0) | 54 (53.5) | |
| Cirrhosis | 0.101 | ||
| No | 14 (11.8) | 7 (5.8) | |
| Yes | 105 (88.2) | 114 (94.2) | |
Bold indicates significant value (P < 0.05).
Abbreviations: AVR- CC, active viral replication chronic carrier; CC, chronic carrier.
TABLE 2.
Uni- and Multivariable Cox Model of Clinical Variables Associated With OS
| Clinical Variable | Hazard Ratio (95% CI) | P Value |
|---|---|---|
|
| ||
| Univariate analysis* | ||
| γδ T-cell-specific genes (LR vs. HR) | 0.57 (0.40–0.83) | 0.003 |
| Age (>50 vs. <50) | 1.11 (0.77–1.60) | 0.567 |
| Sex (female vs. male) | 0.49 (0.26–0.95) | 0.033 |
| HBV (AVR-CC vs. CC) | 1.11 (0.72–1.72) | 0.627 |
| Cirrhosis (yes vs. no) | 3.35 (1.24–9.09) | 0.017 |
| Child- Pugh score (B vs. A) | 1.37 (0.81–2.31) | 0.243 |
| AFP (elevated vs. normal) | 1.35 (0.94–1.95) | 0.103 |
| CA 19–9 (elevated vs. normal) | 1.61 (1.12–2.33) | 0.010 |
| Tumor size (≥3 vs. <3 cm) | 2.20 (1.46–3.31) | <0.001 |
| Multinodular (yes vs. no) | 1.47 (0.97–2.23) | 0.067 |
| Microvascular invasion (yes vs.no) | 1.76 (1.18–2.62) | 0.006 |
| CLIP staging (late vs. early) | 2.34 (1.46–3.77) | <0.001 |
| Okuda staging (1 vs. 0) | 1.62 (0.99–2.64) | 0.051 |
| TNM staging | ||
| I | ref.† | — |
| II | 1.95 (0.69–5.23) | 0.209 |
| III | 3.09 (1.83–5.21) | <0.001 |
| BCLC stage | ||
| 0 | ref. | — |
| A | 3.70 (1.16–11.77) | 0.027 |
| B | 7.83 (2.26–27.14) | 0.001 |
| C | 12.89 (3.84–43.20) | <0.001 |
| Multivariate analysis‡ | ||
| γδ T-cell-specific genes (LR vs. HR) | 0.49 (0.32–0.76) | 0.001 |
| Sex (female vs. male) | 0.53 (0.26–1.11) | 0.091 |
| Cirrhosis (yes vs. no) | 2.06 (0.73–5.83) | 0.175 |
| CA 19–9 (elevated vs. normal) | 2.08 (1.37–3.16) | 0.001 |
| Microvascular invasion (yes vs.no) | 1.46 (0.96–2.20) | 0.074 |
| BCLC stage | ||
| 0 | ref. | — |
| A | 4.80 (1.48–15.56) | 0.009 |
| B | 9.29 (2.58–33.45) | 0.001 |
| C | 14.89 (4.32–51.25) | <0.001 |
| Multivariate analysis§ | ||
| γδ T-cell-specific genes (LR vs. HR) | 0.53 (0.30–0.95) | 0.035 |
| Sex (female vs. male) | 0.38 (0.12–1.18) | 0.094 |
| Cirrhosis (yes vs. no) | 0.45 (0.09–2.24) | 0.330 |
| CA 19–9 (elevated vs. normal) | 1.56 (0.85–2.87) | 0.149 |
| Microvascular invasion (yes vs. no) | 1.60 (0.87–2.94) | 0.133 |
| TNM staging | ||
| I | ref. | — |
| II | 0.81 (0.10–6.43) | 0.842 |
| III | 2.99 (1.61–5.55) | <0.001 |
| Multivariate analysis|| | ||
| γδ T-cell-specific genes (LR vs. HR) | 0.59 (0.38–0.90) | 0.015 |
| Sex (female vs. male) | 0.47 (0.23–0.97) | 0.042 |
| Cirrhosis (yes vs. no) | 1.75 (0.64–4.86) | 0.277 |
| CA 19–9 (elevated vs. normal) | 2.35 (1.14–4.88) | 0.021 |
| Tumor size (≥3 vs. >3 cm) | 1.88 (1.20–2.95) | 0.006 |
| Microvascular invasion (yes vs. no) | 1.46 0.95–2.19) | 0.082 |
| CLIP staging (late vs. early) | 2.35 (1.14–4.88) | 0.021 |
ref = reference variable. Multivariate analysis, Cox proportional hazards regression adjusting for sex, cirrhosis, CA 19–9, tumor size, and microvascular invasion.
BCLC staging.
TNM staging.
CLIP staging.
Bold indicates significant value (P < 0.05).
Abbreviations: AVR-CC,active viral replication chronic carrier; CC, chronic carrier; HR, high risk; LR, low risk.
γδ T-CELL-ASSOCIATED INTRATUMORAL IMMUNE CELL INFILTRATION
γδ T-cell-associated tumors have a higher cytolytic activity and are less aggressive, suggesting the possible roles of γδ T cells to promote an antitumor liver milieu through alteration of immune cell profiles. To test this hypothesis, we performed class comparison analyses between LR and HR subgroups in the LCI cohort of 240 HCC samples using an established 1,622-immune-cell–specific gene set, a strategy used successfully in defining hepatic stellate cell–associated immune cell landscape.(6) By determining the relative fold change between LR and HR, we quantified differentially expressed immune genes specific to certain immune cell types. Accordingly, immune-specific genes most differentially affected between the two subgroups were enriched for lymphoid cell lineage such as T cells, but not the myeloid cell lineage, in the LR subgroup (Fig. 4A). Further analysis by CIBERSORT revealed that although no difference was observed in CD4+ cells (data not shown), CD8+ T cells were elevated and regulatory T (Treg) cells were decreased in the HR group compared to the LR group (Fig. 4B,C). Consistently, expression levels of transcription factors, such as T-box transcription factor 21 (TBX21) and GATA binding protein 3 (GATA3), specific for T-cell linages were significantly higher in the LR group compared to the HR group (Fig. 4D,E). These results suggest that CD8+ T cells might exert a much stronger cytotoxic effect, contributing to a good prognosis of the LR subgroup.
FIG. 4.
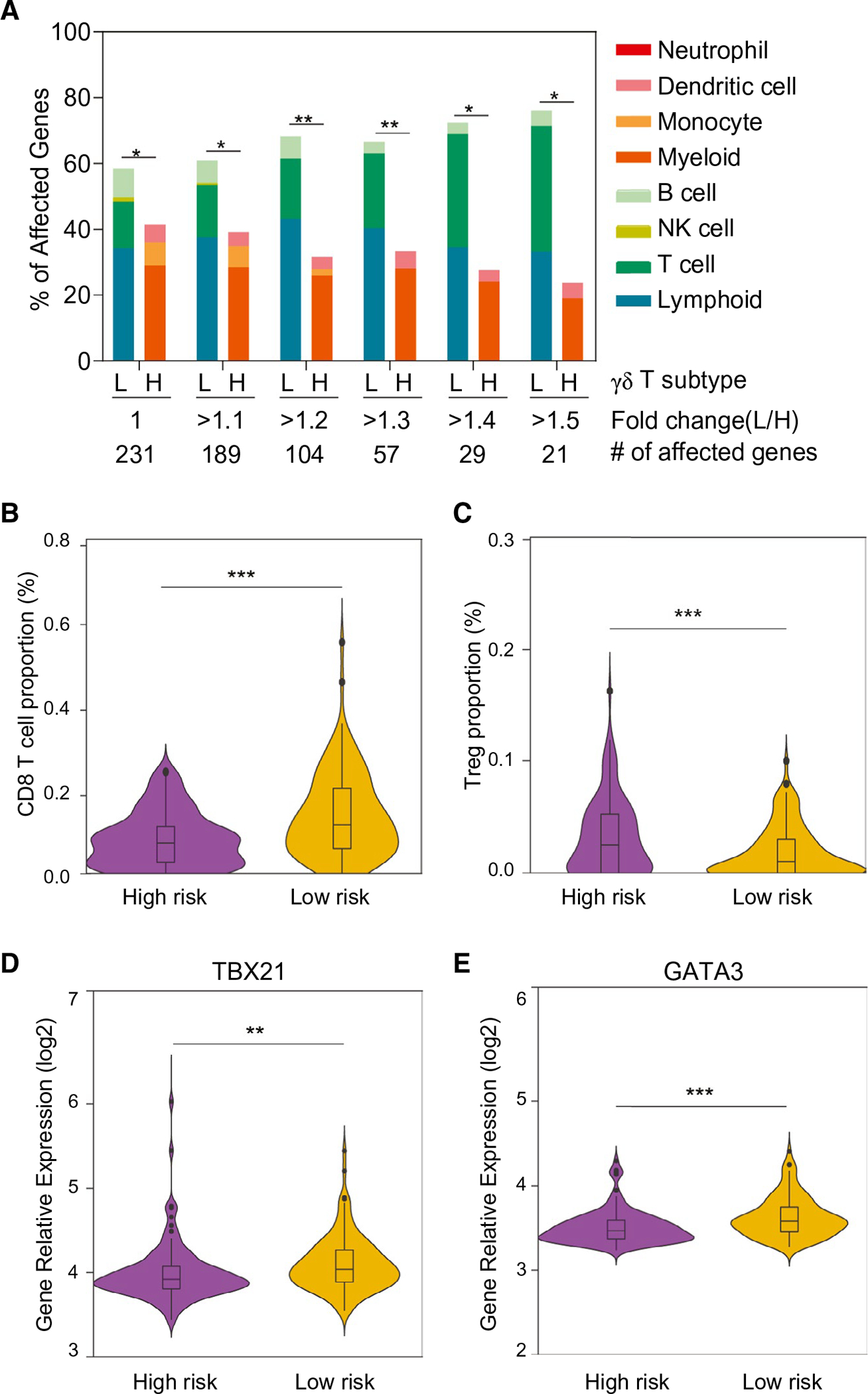
Expression profiles of immune-cell–related genes in the two subgroups defined by γδ T-cell-specific gene signature. (A) Immune-cell gene expression profiles based on γδ T LR and γδ T HR subgroups. Among 1,622 immune cell genes defined by immune response in silico, 231 genes unique for each cell type were used to calculate the percentage of affected genes. Total numbers of significantly expressed genes (adjusted P < 0.05) with different fold changes between the LR and HR subgroups are indicated. *P < 0.05; **P < 0.01 from the hypergeometric probability test. (B,C) The composition of two main immune cells’ subgroups between the predefined low- and high-risk HCC tumor groups. Proportion of CD8+ (B) and Treg (C) were calculated using the array data. Comparison of the expression levels of the transcription factors, TBX21 (D) and GATA3 (E), involved in the T-cell differentiation between the predefined low- and high-risk HCC tumor groups. Abbreviation: L/H, low/high. **P < 0.001; ***P < 0.001.
RECRUITMENT OF γδ T CELLS TO THE TUMOR MICROENVIRONMENT
It was noted that although the levels of γδ T cells were significantly higher in nontumor tissues than paired tumor tissues, tumors from the LR group had significantly more intratumoral γδ T-cell infiltrates than that of the HR group. These results suggest a difference in the recruitment of intratumoral γδ T cells among HCC cases. To determine what contributes to the differential levels of intratumoral γδ T cells, we performed class comparison analysis of tumor transcriptome data between HR and LR groups in the LCI cohort. We found 974 differentially expressed genes (univariate, P < 0.001; Fig. 5A). Among them, 160 genes had >1.5-fold differences (false discovery rate, <0.05). We then performed gene set enrichment analysis (GSEA; Supporting Table S2), and found that genes involved in the chemokine pathway are significantly enriched among the differential expression gene set (Fig. 5B). To further identify specific targets highly correlated with γδ T-cell activity, we performed correlation analysis between levels of γδ T cells in each tumor based on the weight of the expression of γδ T-cell-associated genes (Supporting Table S3) and known chemokines (n = 37) and their corresponding receptors (n = 19). We found that the chemokine (C-C motif ) ligand 4 (CCL4) and CCL5 were the top two chemokines positively correlated with γδ T cells (Fig. 5C; Supporting Table S4). Encouragingly, CCR5, a receptor shared by CCL4/CCL5, was the top chemokine receptor positively correlated with γδ T cells (Supporting Table S4; Fig. 5C). Consistently, expression levels of CCL4, CCL5, and CCR5 in the HR group were much lower than that of the LR groups (Fig. 5D). Similar results were observed in the TCGA cohort (Supporting Table S5; Supporting Fig. S2). Collectively, these results suggest that CCL4/5 and their receptor, CCR5, may play an important role in the recruitment of γδ T cells into the tumor microenvironment.
FIG. 5.
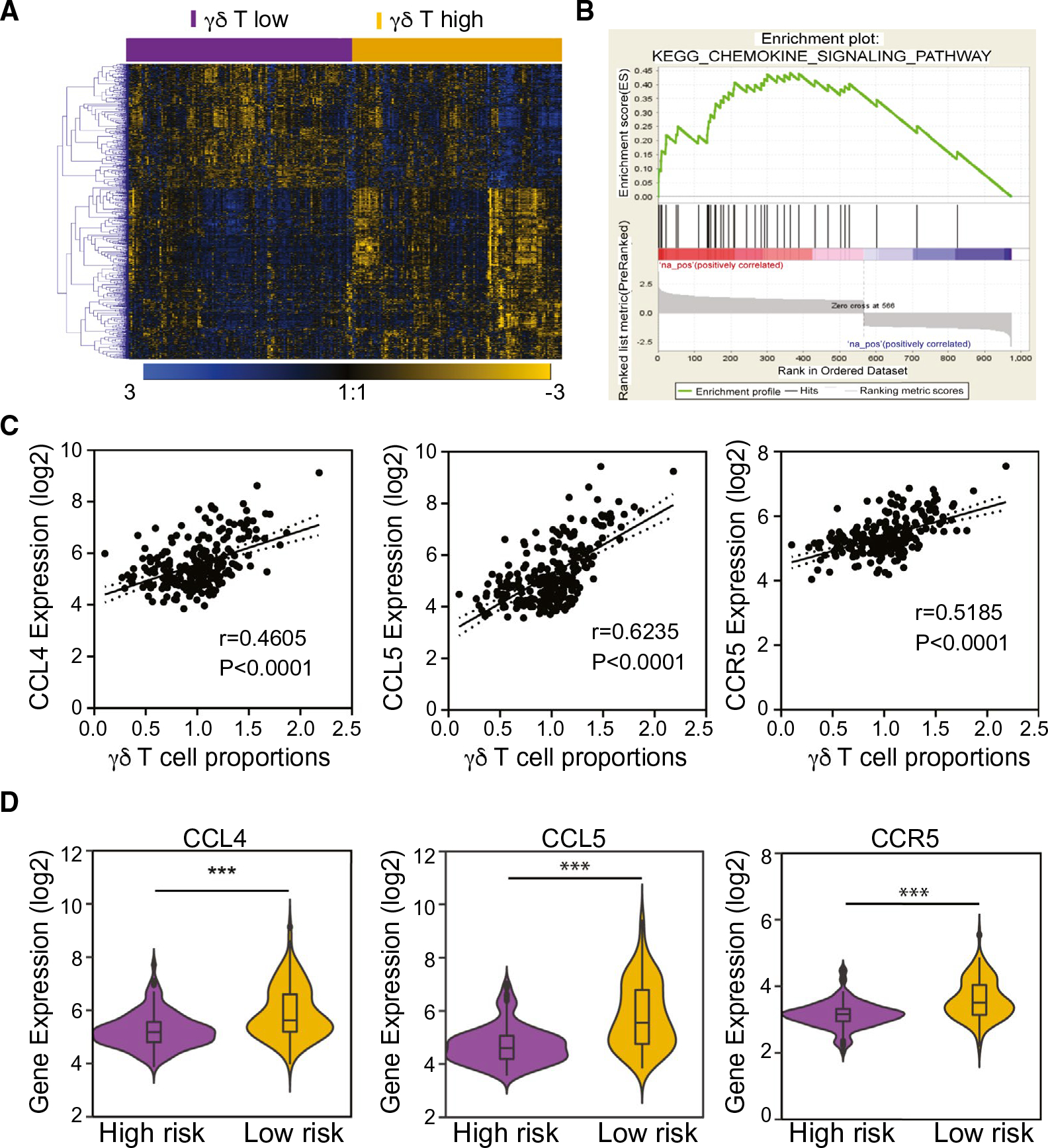
Gene expression profiles of HCC tumor tissues stratified by γδ T-cell gene signature predicted low- and high-risk subgroups. (A) Hierarchical clustering of the 974 differentially expressed genes (P < 0.001) between γδ T-cell-signature–predicted subgroups. Pseudocolors indicate transcript levels above (blue), below (yellow), or equal to (black) the mean, respectively. Genes were ordered by centered correlation and complete linkage. The scale represents gene expression level from −3.0 to 3.0 in a log2 scale. (B) Result of GSEA enrichment of chemokine signal pathway based on the genes highly expressed in the γδ T high-tumor subgroup. (C) Pearson’s correlation analysis between CCL4, CCL5, and CCR5 expression level and γδ T-cell weights in HCC tumor tissues from the LCI cohorts. (D) Comparison of expression of CCL4, CCL5, and CCR5 in the subgroups defined by γδ T-cell signature in tumor tissues from the LCI cohorts. Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes. ***P < 0.001.
To further validate the above results, we performed IHC staining of anti-CCL5 in FFPE liver cancer tissues in the LCI cohort. The results indicated that CCL5 is mainly expressed by the infiltrated lymphocytes in the nontumor tissues (Supporting Fig. S3, left panel), whereas it could be expressed by both the TILs and tumor cells in tumor tissues (Supporting Fig. S3, right panel). Furthermore, we also found that CCL5 expression level correlated significantly with the number of TCRγδ-positive staining cells (r = 0.5993; P < 0.0001; Fig. 6A). CCL5 expression level was significantly higher in tumor samples in which more γδ T cells infiltrated (Fig. 6B). These data suggest that the interaction between CCL4/CCL5 and CCR5 played an important role in HCC’s tumor microenvironment, which may be the main contributor for γδ T-cell migration from peritumor tissues to the tumor regions.
FIG. 6.
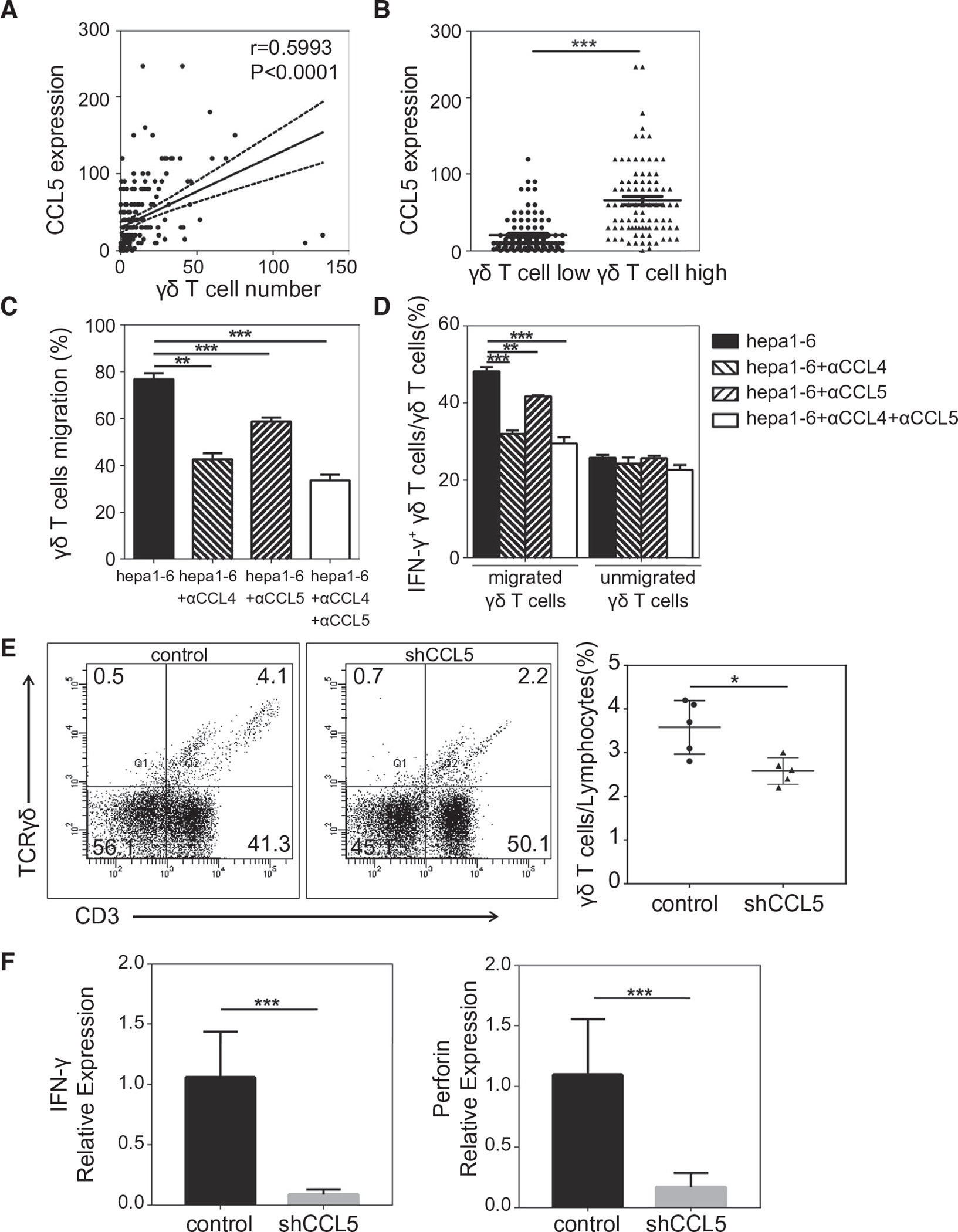
Roles of tumor-cell–derived CCL4/5 on γδ T cells. (A) Spearman’s correlation analysis between CCL5 expression level and TCRγδ-positive staining cells number in the 182 FFPE HCC tissues. (B) Comparison of CCL5 expression levels between the high and low γδ T-cell-infiltrating groups. (C) Results of transwell migration assay, reflected by the proportion of chemo-attracted γδ T cells by murine hepatoma cell line Hepa1–6 with or without neutralized anti-CCL4/5. (D) Comparison of IFN-γ expression levels in transmigrated and unmigrated γδ T cells in the coculture model with Hepa1–6. (E,F) B6 mice were treated with intrahepatic trasplantation of either CCL5 shRNA-transduced or control oligo-transduced Hepa 1–6. At day 14, tumor-bearing livers were excised and used for the analysis of γδ T-cell proportion by flow cytometry (E) or analysis of expression levels of IFN-γ and perforin by real-time RT-PCR (F). Abbreviation: shCCL5, chemokine (C-C motif) ligand 5 with short hairpin RNA. *P < 0.05; **P < 0.01; ***P < 0.001.
We used an syngeneic orthotopic murine HCC model to determine a functional link between CCL4/CCL5 and γδ T cells. We first determined the CCR5 expression patterns in murine spleen and liver lymphocytes with flow cytometry and found that the expression level of CCR5 on γδ T cells was significantly greater than CD4+ T and CD8+ T cells (Supporting Fig S4A). We next confirmed that murine hepatoma cell line Hepa1–6 expressed both CCL4 and CCL5 (Supporting Fig S4B). We then investigated whether tumor-cell–derived CCL4/CCL5 could promote γδ T-cell activation and recruitment in vitro. Transwell migration assay revealed that γδ T cells were recruited by Hepa1–6 efficiently, and this trafficking could be neutralized by anti-CCL4/5 antibody (αCCL4/5), indicating that Hepa1–6-derived CCL4/5 may play vital roles in the chemotaxis of γδ T cells (Fig 6C). Importantly, antibody neutralization of CCL4/5 also decreased the secretion potency of IFN-γ, a major cytokine produced by γδ T cells in antitumor immunity (Fig 6D and Supporting Fig S5A). To further explore the chemotactic activity of tumor-cell–derived CCL5 in vivo, an orthotopic mouse HCC model was used. We tested three CCL5 shRNA oligos (i.e., shRNA1, shRNA2, and shRNA3), and found that shRNA3 was most efficient in reducing levels of CCL5 in Hepa1–6 cells (Supporting Fig. S5B). Strikingly, levels of γδ T cells were significantly reduced in mouse livers transplanted with shRNA3-transduced Hepa1–6 cells compared to control oligos-transduced Hepa1–6 cells (Fig. 6E). Consistently, expression levels of IFN-γ and perforin were also significantly reduced upon CCL5 silencing (Fig. 6F). Taken together, these results indicate that tumor-cell–produced CCL4/5 may serve as chemokines for γδ T cells.
Discussion
The immune landscape of the tumor microenvironment can influence cancer initiation, progression, and invasion that contribute to patient outcomes.(25,26) The liver, as an important immunological organ, is enriched with macrophages (Kupffer cells), natural killer cells (NK), and natural killer T (NKT) cells and is among the richest sources for γδ T cells in the body.(27) It is now well established that γδ T cells possess cytotoxic antitumor activity mediated by the production of proinflammatory cytokines, direct cytotoxic activity, and regulation of the biological functions of other cell types.(28) Recent studies have demonstrated that low proportions or functional impairment of γδ T cells correlate with tumor progression and recurrence after curative resection,(29,30) consistent with our findings.(7) In this study, we established a γδ T-cell gene signature to explore their biological roles in HCC. We show that the γδ T-cell gene signature is predictive of HCC recurrence and OS. In measuring the transcript levels of GZMA and PRF1, we demonstrate that HCC patients in the HR group had a lower cytolytic activity than the LR group. These findings were also validated in the TCGA cohort and confirmed using IHC analyses. Together, our results suggest that there are more γδ T-cell infiltration and stronger cytotoxicity of γδ T cells in the LR group compared to the HR HCC subgroup, indicative of a higher antitumor immune status.
NKG2D, a well-studied NK cell receptor predominantly expressed by γδ T cells, plays an important role in cytotoxicity of tumor cells. That DNAM-1 and CD96 (tactile) engagement could induce IFN-γ production by γδ T cells has been reported to enhance the HCC tumor cell lysis induced by γδ T cells.(24) By measuring the cytolytic activity and expression levels of three NK cell receptors to evaluate the cytotoxic function of γδ T cells, we found that both the number and ability of tumor-cell lytic activity of γδ T cells were significantly correlated with γδ T-cell-specific gene expression, which was also validated in the TCGA cohort. Above all, these results support the hypothesis that γδ T cells play a protective role in the progression and prognosis of HCC. In addition, our clinical data analyses further supported the notion that HCC patients with a higher γδ T-cell proportion, such as the LR subgroup, had a better OS and a less aggressive tumor type. These patients overall had less active viral replication, present with less tumor numbers, less microvascular invasion, and, subsequently, an earlier tumor stage. Multivariate analyses including various clinical risk factors and various clinical staging indicated that the γδ T-cell gene signature was an independent predictor of survival, making it a useful tool to predict intratumor infiltration of γδ T cells and HCC patients’ prognosis.
Chemokines are low-molecular-weight cytokines that control leukocyte trafficking in inflammation and homeostasis, modulate immune responses, and have important functions in embryonic development, hematopoiesis, angiogenesis, and metastasis.(31) Our analysis between γδ T-cell expression and the 37 chemokines and 19 chemokine receptors demonstrate that CCL4 and CCL5 were the top two chemokines positively correlated with γδ T-cell expression in both the LCI and TCGA cohorts. CCL4, also known as macrophage inflammatory protein-1 β, is a chemoattractant for NK cells, monocytes, and a variety of other immune cells.(32) CCL5 has a strong chemotactic activity toward multiple immune cells and plays an active role in recruiting leukocytes into inflammatory sites.(33) Together, CCL4 and CCL5 show specificity for CCR5 receptors. CCR5 is expressed by various subsets of activated T lymphocytes, including CD4+ T helper 1 cells, CD8+ cytotoxic T cells, Treg cells, NK and NKT cells, monocytes, macrophages, and also γδ T cells.(31) Through checking the expression of CCL4, CCL5, and CCR5 in tumor tissues, we found that their expression level was significantly increased in the LR subgroup. Similarly, IHC analyses showed similar trends and a positive correlation, suggesting that CCL4 and CCL5 may be the triggering factor of γδ T-cell accumulation in the HCC tumor microenvironment. Furthermore, in vitro and in vivo experiments have validated CCL4 and CCL5 as chemokines for γδ T cells.
In summary, the current study revealed that the γδ T-cell gene signature can predict the OS and recurrence-fr ee survival of patients with HCC. Increasing infiltrated γδ T cells may play protective roles by regulating the infiltration and differentiation of CD8+ T cells in HCC development. Chemokines, such as CCL4 and CCL5, may play a triggering role in the accumulation of γδ T cells. However, what regulates the expression of CCL4/5 in tumor cells and how they induce tumor-infiltrating γδ T cells to be antitumorigenic remain open for further investigation. For example, γδ T cells could be divided into different subtypes whereas only a specific γδ T-cell type may be more effective in attacking cancer cells. Functional polarization of γδ T cells could be determined by analyzing the composition and CCR5 expression levels of different subtypes of γδ T cells. Moreover, the antitumor effects of γδ T cells could also be affected by other cellular factors within the tumor microenvironment. Signals from other tumor-infiltrating immune cells can affect the cytokine production, cytotoxicity, and exhaustive status of the local resident or migrated γδ T cells. Various cues from the tumor microenvironment, including oxygen tension, nutrient availability, and cancer cell metabolites, may also regulate antitumor γδ T-cell functions. Chemotherapeutic agents could also regulate the activity of γδ T cells. These are worthy research topics for further exploitation in order to determine the exact mechanism underling the differentiation and homeostasis of γδ T cells. Further efforts may be needed to determine the relationship between the tumor microenvironment and changes of CCR5 expression in the context of γδ T-cell migration. Mechanistically modulating intratumoral infiltration of γδ T cells, thereby altering the composition of the tumor immune microenvironment, could be exploited as a viable strategy to improve HCC therapeutic intervention.
Supplementary Material
Acknowledgments
Supported by the Intramural Research Program of the Center for Cancer Research of the National Cancer Institute (ZIA-BC010313, ZIA BC 011870). N.Z. was supported by the Program of Study Abroad for Young Scholar sponsored by the China Scholarship Council (No. 201706940010).
Abbreviations:
- AFP
alpha-fetoprotein
- BCLC
Barcelona Clinic Liver Cancer
- CA 19-9
cancer antigen 19-9
- CCL4
chemokine (C-C motif) ligand 4
- CCL5
chemokine (C-C motif) ligand 5
- CCR5
C-C chemokine receptor type 5
- CLIP
Cancer Liver Italian Program
- DNAM-1
DNAX accessory molecule 1
- FFPE
paraffin-embedded, formalin-fixed
- GATA3
GATA binding protein 3
- GZMA
granzyme A
- H&E
hematoxylin and eosin
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HR
high-risk group
- IFN
interferon
- IHC
immunohistochemical
- KLRK1
killer cell lectin-like receptor K1
- LCI
Liver Cancer Institute
- LR
low-risk group
- NK
natural killer cells
- NKT
natural killer T
- OKUDA
Okuda staging system
- OS
overall survival
- PRF1
perforin 1
- RANTES
regulated on activation, normal T expressed and secreted
- shRNA
short hairpin RNA
- TBX21
T-box transcription factor 21
- TCGA
The Cancer Genome Atlas
- TCR
T-cell receptor
- TILs
tumor-infiltrating lymphocytes
- TMA
tissue microarray
- TNM
Tumor Node Metastasis
Footnotes
Potential conflict of interest: Nothing to report.
Supporting Information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.31412/suppinfo.
REFERENCES
- 1).Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2).Mao B, Wang G. MicroRNAs involved with hepatocellular carcinoma (Review). Oncol Rep 2015;34:2811–2820. [DOI] [PubMed] [Google Scholar]
- 3).Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010;140:883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Budhu A, Wang XW. The role of cytokines in hepatocellular carcinoma. J Leukoc Biol 2006;80:1197–1213. [DOI] [PubMed] [Google Scholar]
- 5).Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell 2006;10:99–111. [DOI] [PubMed] [Google Scholar]
- 6).Ji J, Eggert T, Budhu A, Forgues M, Takai A, Dang H, et al. Hepatic stellate cell and monocyte interaction contributes to poor prognosis in hepatocellular carcinoma. Hepatology 2015;62:481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM, et al. Common molecular subtypes among asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell 2017;32:57–70.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Gaud G, Lesourne R, Love PE. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol 2018;18:485–497. [DOI] [PubMed] [Google Scholar]
- 9).Chien YH, Bonneville M. Gamma delta T cell receptors. Cell Mol Life Sci 2006;63:2089–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Rajoriya N, Fergusson JR, Leithead JA, Klenerman P. Gamma delta T-lymphocytes in hepatitis C and chronic liver disease. Front Immunol 2014;5:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Kasper HU, Ligum D, Cucus J, Stippel DL, Dienes HP, Drebber U. Liver distribution of gammadelta-T-cells in patients with chronic hepatitis of different etiology. APMIS 2009;117:779–785. [DOI] [PubMed] [Google Scholar]
- 12).Martinez-Llordella M, Puig-Pey I, Orlando G, Ramoni M, Tisone G, Rimola A, et al. Multiparameter immune profiling of operational tolerance in liver transplantation. Am J Transplant 2007;7:309–319. [DOI] [PubMed] [Google Scholar]
- 13).Koshiba T, Li Y, Takemura M, Wu Y, Sakaguchi S, Minato N, et al. Clinical, immunological, and pathological aspects of operational tolerance after pediatric living-donor liver transplantation. Transpl Immunol 2007;17:94–97. [DOI] [PubMed] [Google Scholar]
- 14).Puig-Pey I, Bohne F, Benitez C, Lopez M, Martinez-Llordella M, Oppenheimer F, et al. Characterization of gammadelta T cell subsets in organ transplantation. Transpl Int 2010;23:1045–1055. [DOI] [PubMed] [Google Scholar]
- 15).Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Can Res 2010;70:10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).The Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327–1341.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Brown JP, Chandra A. Science made simple: tissue microarrays (TMAs). BJU Int 2014;114:294–295. [DOI] [PubMed] [Google Scholar]
- 18).Choi CH, Chung JY, Kim JH, Kim BG, Hewitt SM. Expression of fibroblast growth factor receptor family members is associated with prognosis in early stage cervical cancer patients. J Transl Med 2016;14:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).He W, Hao J, Dong S, Gao Y, Tao J, Chi H, et al. Naturally activated V gamma 4 gamma delta T cells play a protective role in tumor immunity through expression of eomesodermin. J Immunol 2010;185:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Yao X, Hu JF, Daniels M, Yien H, Lu H, Sharan H, et al. A novel orthotopic tumor model to study growth factors and oncogenes in hepatocarcinogenesis. Clin Cancer Res 2003;9:2719–2726. [PubMed] [Google Scholar]
- 21).Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Duseja A. Staging of hepatocellular carcinoma. J Clin Exp Hepatol 2014;4(Suppl. 3):S74–S79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Toutirais O, Cabillic F, Le Friec G, Salot S, Loyer P, Le Gallo M, et al. DNAX accessory molecule-1 (CD226) promotes human hepatocellular carcinoma cell lysis by Vgamma9Vdelta2 T cells. Eur J Immunol 2009;39:1361–1368. [DOI] [PubMed] [Google Scholar]
- 25).De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer 2017;17:457–474. [DOI] [PubMed] [Google Scholar]
- 26).Maman S, Witz IP. A history of exploring cancer in context. Nat Rev Cancer 2018;18:359–376. [DOI] [PubMed] [Google Scholar]
- 27).Hammerich L, Tacke F. Role of gamma-delta T cells in liver inflammation and fibrosis. World J Gastrointest Pathophysiol 2014;5:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Nielsen MM, Witherden DA, Havran WL. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol 2017;17:733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Cai XY, Wang JX, Yi Y, He HW, Ni XC, Zhou J, et al. Low counts of gammadelta T cells in peritumoral liver tissue are related to more frequent recurrence in patients with hepatocellular carcinoma after curative resection. Asian Pac J Cancer Prev 2014;15:775–780. [DOI] [PubMed] [Google Scholar]
- 30).Yi Y, He HW, Wang JX, Cai XY, Li YW, Zhou J, et al. The functional impairment of HCC-infiltrating γδ T cells, partially mediated by regulatory T cells in a TGFbeta- and IL-10-dependent manner. J Hepatol 2013;58:977–983. [DOI] [PubMed] [Google Scholar]
- 31).Gonzalez-Martin A, Mira E, Manes S. CCR5 as a potential target in cancer therapy: inhibition or stimulation? Anticancer Agents Med Chem 2012;12:1045–1057. [DOI] [PubMed] [Google Scholar]
- 32).Cheng NL, Chen X, Kim J, Shi AH, Nguyen C, Wersto R, et al. MicroRNA-125b modulates inflammatory chemokine CCL4 expression in immune cells and its reduction causes CCL4 increase with age. Aging Cell 2015;14:200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Lapteva N, Huang XF. CCL5 as an adjuvant for cancer immunotherapy. Expert Opin Biol Ther 2010;10:725–733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


