Figure 1.
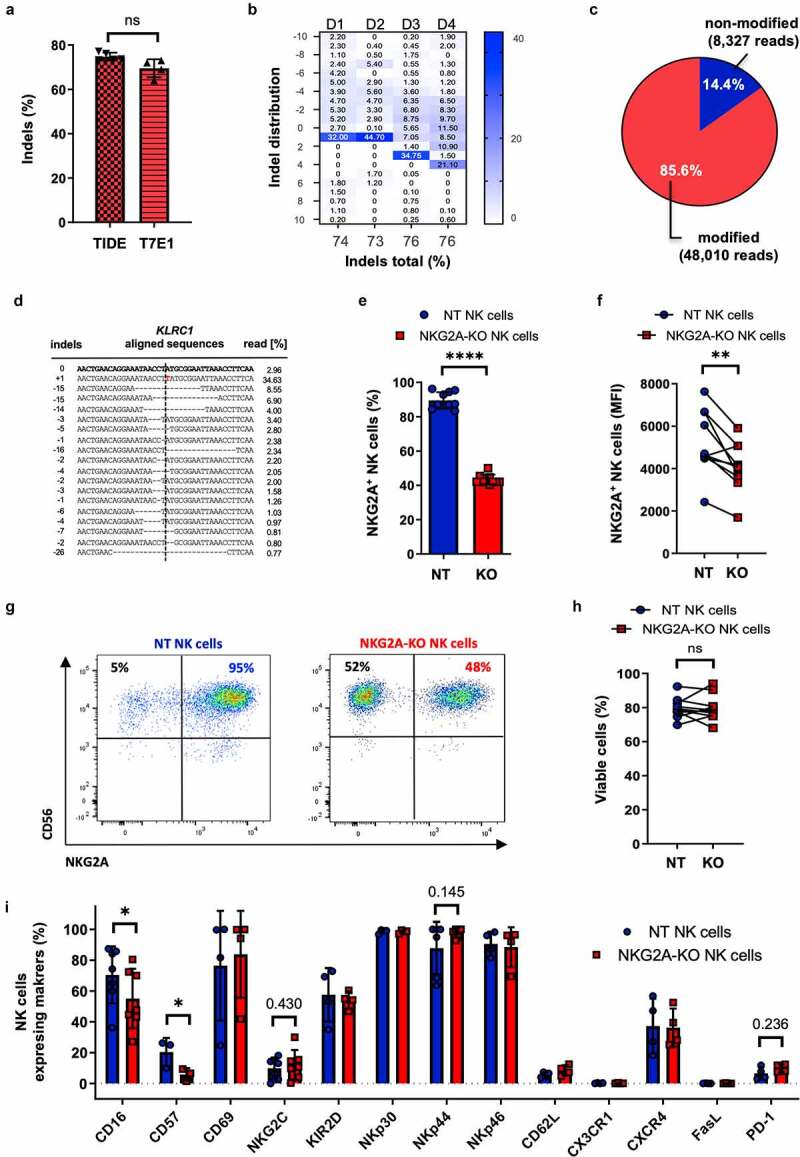
Characterization of KLRC1-edited primary NK cells. Primary NK cells were nucleofected with CRISPR-Cas9 ribonucleoprotein (RNP) complex targeting KLRC1 locus. (a-d) Genotyping. Frequency of KLRC1 disruption was evaluated by T7E1 assay (a), TIDE analysis (a, b), and targeted amplicon next-generation sequencing (NGS) (c-d). Insertion/deletion (indel) distribution profiles were analyzed by TIDE for four different donors (D1-4) while NGS was performed for a single donor. (e-i) Phenotyping. KLRC1-edited NKG2A knock-out NK cells (NKG2A-KO NK cells) were compared to non-treated NK cells (NT NK cells) by flow cytometry (n = 10 donors). Percentages of NKG2A-positive NK cells (e), Mean Fluorescence Intensities (MFI) (f), FACS plots of a representative donor (g) and percentages of viabilities (h) are indicated. Analyses of NK cell receptor expression was measured by flow cytometry (n = 4 donors) (i). Paired t-test. **** p ≤ .0001; ** p ≤ .01; * p ≤ .05; ns p > .05; if not mentioned, no significant difference was observed. T7E1, T7 endonuclease I; TIDE, tracking of indels by decomposition.
