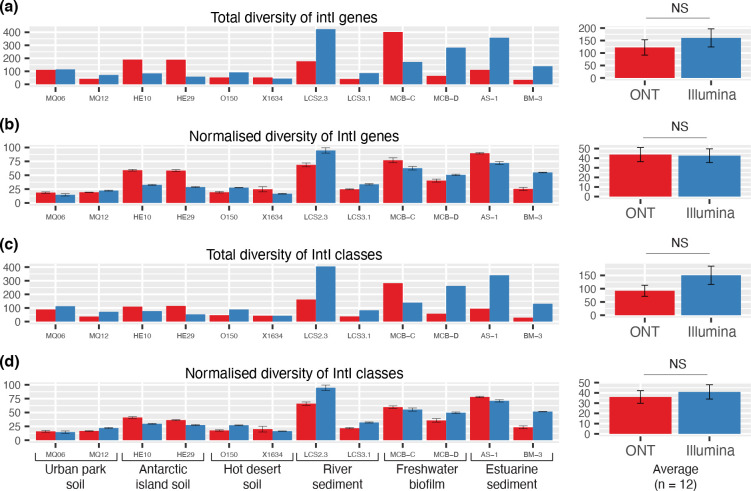
Fig. 3.
Diversity of integron integrases recovered by the intI-R / HS286 primer set. (a) Total non-redundant (100 % amino acid identity) integron integrases (IntIs) recovered. (b) IntI diversity was normalised for sequencing, based on averages (±1 S.E) of triplicate 50-megabase subsamples of raw sequence reads. Total (c) and normalised (d) diversity of IntI classes (using a 94 % amino acid clustering threshold) are shown. Average (±1 S.E) diversity for each analysis are shown on the right-hand side of each panel. Differences between Nanopore (ONT) and Illumina MiSeq technologies were not significant (NS) as determined by Wilcoxon rank sum tests.