Figure 3 |. RdRp template switching at sites of sequence complementarity models rare events in SARS-CoV-2.
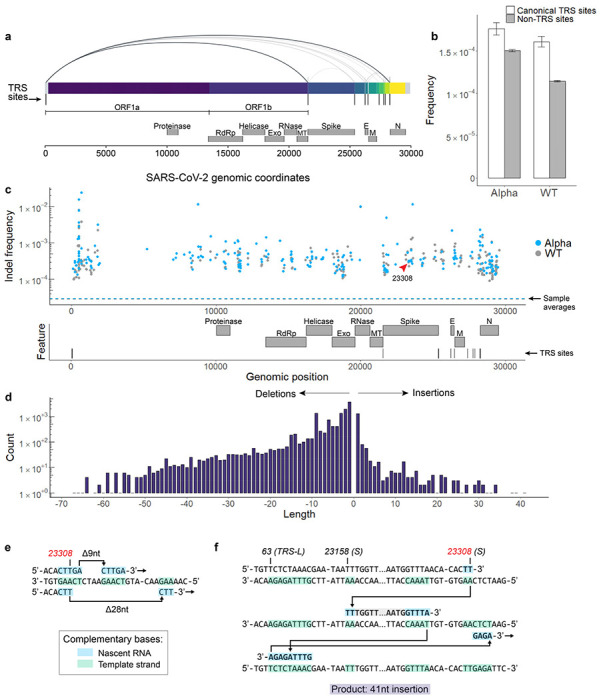
a, Using a spliced aligner (STAR) for mapping, chimeric reads are detected in WT virus. Recurrent jumps between canonical TRSs are visualized as arcs connecting the 5’ and 3’ ends of each chimeric junction. These jumps signify programmed RdRp template switching that functions in viral gene expression. b, TRS regions had a higher RNA variant frequency compared to control regions in both WT and Alpha, suggesting that programmed polymerase jumping reduces overall fidelity in these regions. c, Among other low-fidelity regions are indel hot spots, or loci with significantly elevated frequencies of insertions and deletions. Indel hot spots are calculated by Fisher exact test, filtered for ≥10,000X depth, and graphed by position for both Alpha and WT. d, The size spectrum of insertions and deletions in WT virus reveals rare, large events, many of which appear templated from within the SARS-CoV-2 genome. e, f, Templated indels can be explained by non-programmed RdRp jumping and realignment at sites of sequence complementarity outside of canonical TRSs. Three events from tARC-seq data are modeled, all occurring at the same indel hot spot in the S gene (g.23308, indicated by the red arrow in panel c). The full sequence of the 41-nt insertion (f) is: TGGTTAAAAACAAATGTGTCAATTTCAACTTCAATGGTTTA.
