Figure 4 |. On the origin of Omicron.
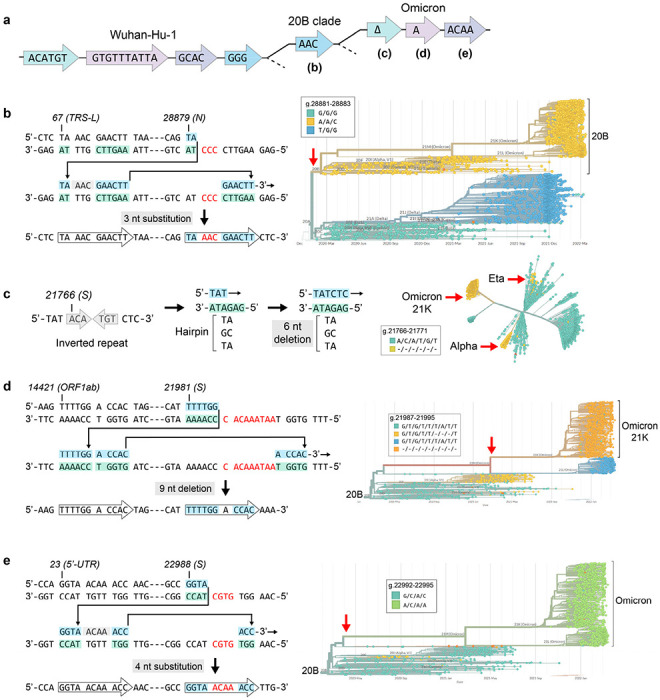
Pandemic data shows that many complex mutations in SARS-CoV-2 appeared suddenly (red arrows). They likely did not accumulate gradually but were driven by a single event: RdRp template switching. In the events modeled here, 3’ complementarity facilitates the misalignment and realignment of RdRp, creating complex mutations that have fueled viral evolution. a, Phylogenetic tree based on sequence alterations that define the 20B and Omicron clades; not drawn to any scale. Discrete, coordinated nucleotide alterations are coded by color, and each template switching event is mapped out below (b-e). b, A GGG>AAC mutation in the N gene occurred once early in the pandemic and helped define the 20B clade. c, A small hairpin in S has spawned the same 6-nt deletion on more than 3 separate occasions, while other single events are specific to Omicron (d-e). Phylogenetic trees were constructed in Nextstrain v2.35.032 from genomes sequenced between Dec. 2019 and March 2022.
