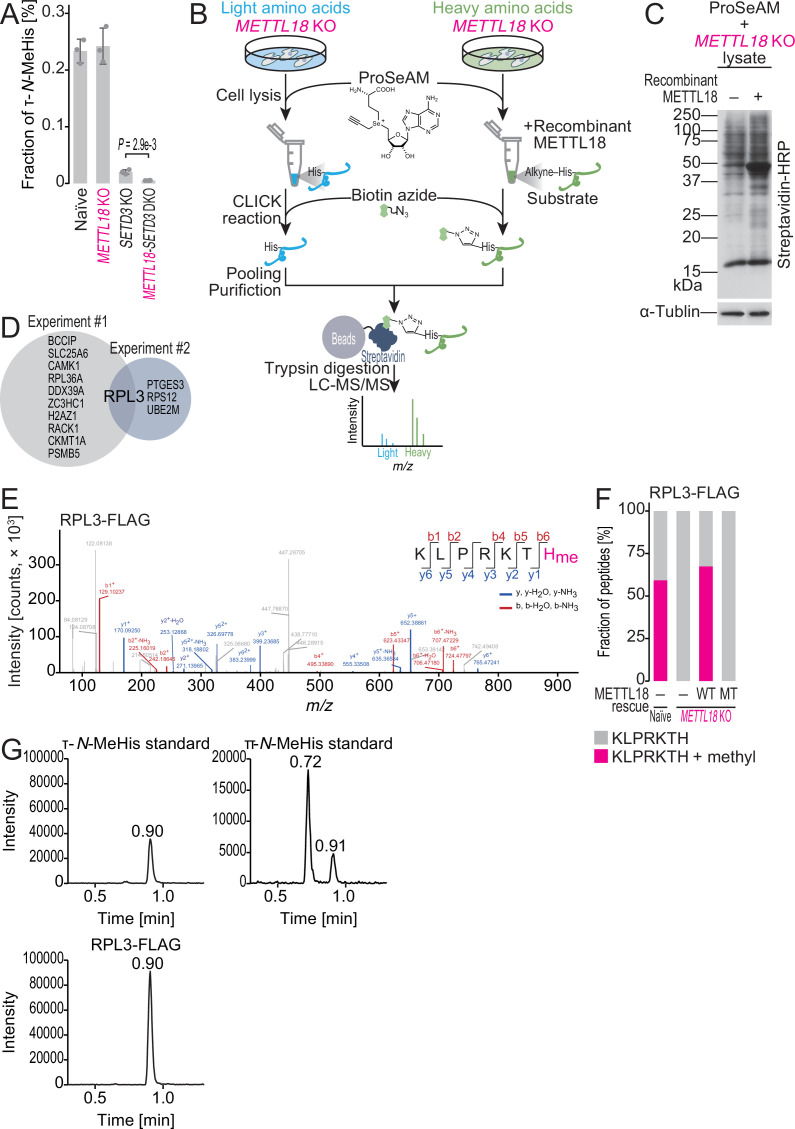
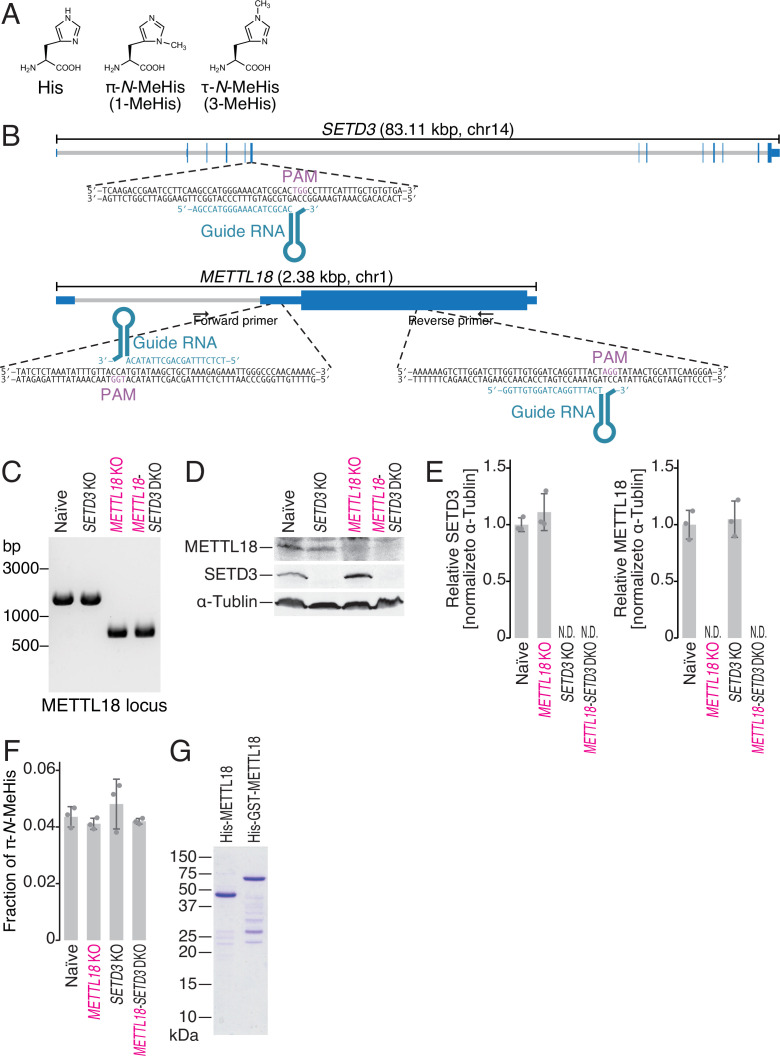
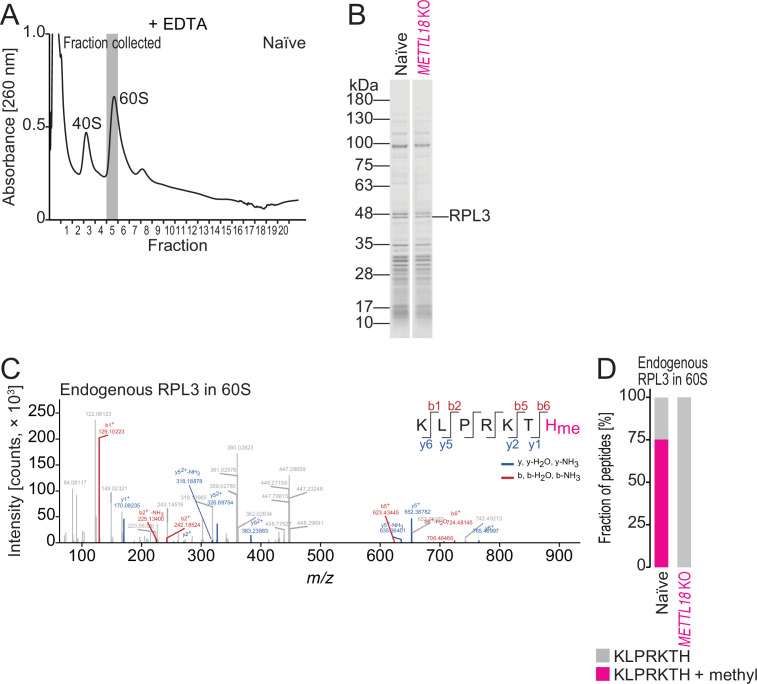
Figure 1. ProSeAM-SILAC identifies RPL3 as a substrate of METTL18.
(A) Multiple reaction monitoring (MRM)-based identification of τ-N-methylated histidine in bulk proteins from the indicated cell lines. Data from three replicates (points) and the mean (bar) with SD (error bar) are shown. Significance was determined by Student’s t-test (unpaired, two-sided). (B) Schematic representation of the ProSeAM-SILAC approach. (C) ProSeAM-labeled proteins in cell lysate with recombinant His-METTL18 protein. Biotinylated proteins were detected by streptavidin-HRP. Western blot for α-tubulin was used as a loading control. (D) Venn diagram of proteins identified in two independent ProSeAM-SILAC experiments. The reproducibly detected protein was RPL3. (E) Methylated histidine residue in ectopically expressed RPL3-FLAG was searched by liquid chromatography mass spectrometry (LC-MS/MS). (F) Quantification of methylated and unmethylated peptides (KLPRKTH) from the indicated cells. RPL3-FLAG was ectopically expressed and immunopurified for LC-MS/MS. WT, wild type; MT, Asp193Lys-Gly195Arg-Gly197Arg mutant. (G) MRM-based identification of τ-N-methylhistidine in peptides from RPL3. The τ-N-methylhistidine standard, π-N-methylhistidine standard, and RPL3-FLAG peptide (KLPRKTH) results are shown. MeHis, methylhistidine.