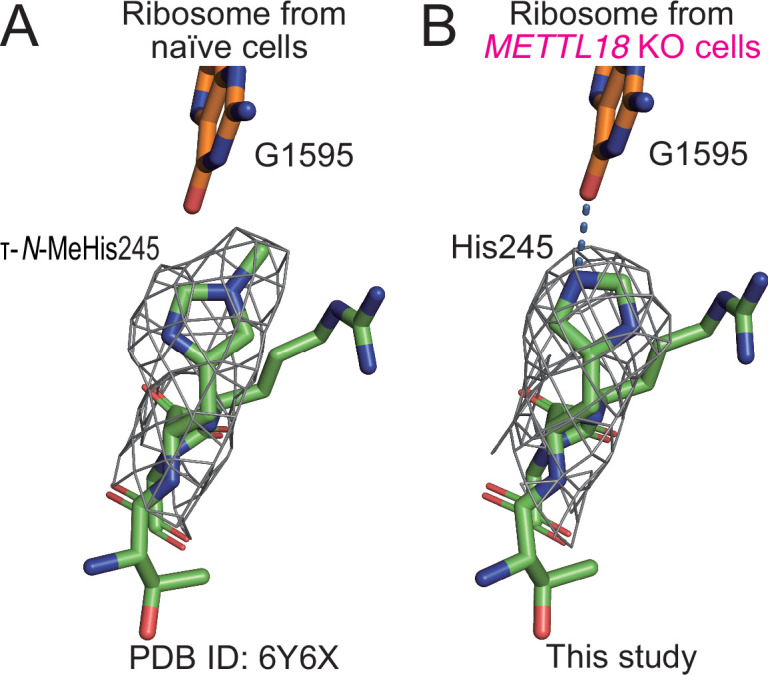
Figure 3. Structural differences in ribosomes upon methylation at His245.
(A) Stick models of 244GHR246 of RPL3 and G1595 of the 28S rRNA of the human ribosome are shown with the cryo-electron microscopy (cryo-EM) density map around His245. The τ-N-methyl group was manually added to the original model (PDB ID: 6Y6X) (Osterman et al., 2020) based on the cryo-EM density map. (B) The same model as in (A) of human ribosome from METTL18 knockout (KO) cells. A hydrogen bond between His245 and G1595 is indicated with a dotted blue line.