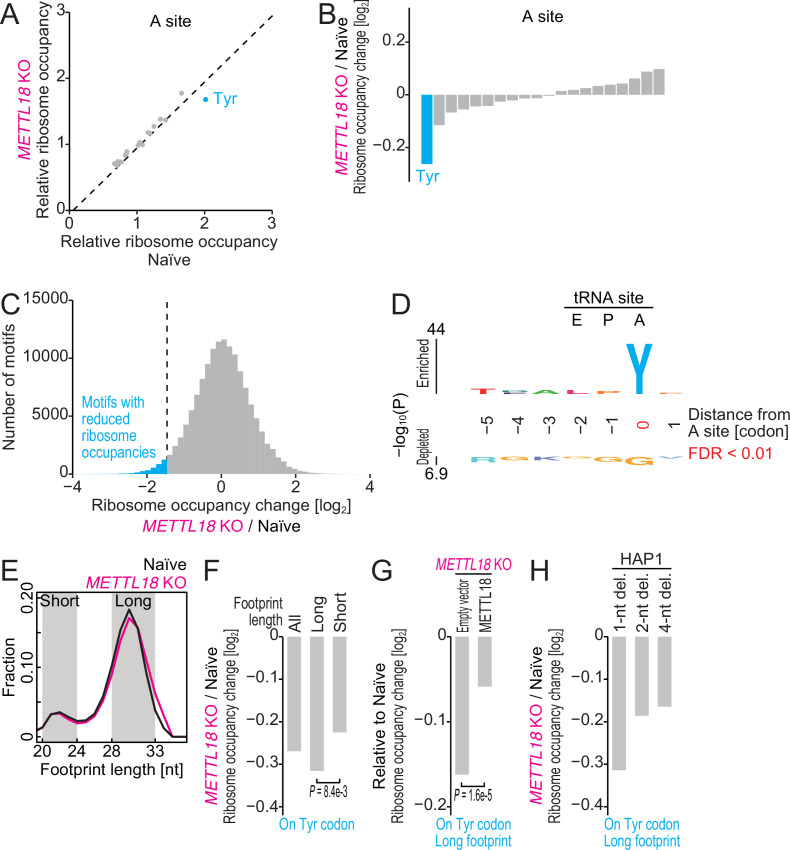
Figure 4. Ribosome profiling reveals Tyr codon-specific translation retardation by RPL3 methylation.
(A) Ribosome occupancy at A-site codons in naïve and METTL18 knockout (KO) HEK293T cells. Data were aggregated into codons with each amino acid species. (B) Ribosome occupancy changes at A-site codons caused by METTL18 KO. (C) Histogram of ribosome occupancy changes in METTL18 KO cells across motifs around A-site codons (seven amino acid motifs). Cyan: motifs with reduced ribosome occupancy (defined by ≤ mean – 2 SD). (D) Amino acid motifs associated with reduced ribosome occupancy in METTL18 KO cells (defined in C) are shown relative to the A-site (at the 0 position). (E) Distribution of footprint length in naïve and METTL18 KO HEK293T cells. (F) Ribosome occupancy changes on Tyr codons by METTL18 KO along all, long (28–33 nt), and short (20–24 nt) footprints. Significance was determined by the Mann–Whitney U-test. (G) The recovery of long footprint reduction in METTL18 KO cells by ectopic expression of METTL18 protein. Significance was determined by the Mann–Whitney U-test. (H) Changes in ribosome occupancy on Tyr codons by METTL18 KO in HAP1 cells along long (28–33 nt) footprints. Del., deletion. In (A–C) and (E–H), the means of two independent experiments are shown.