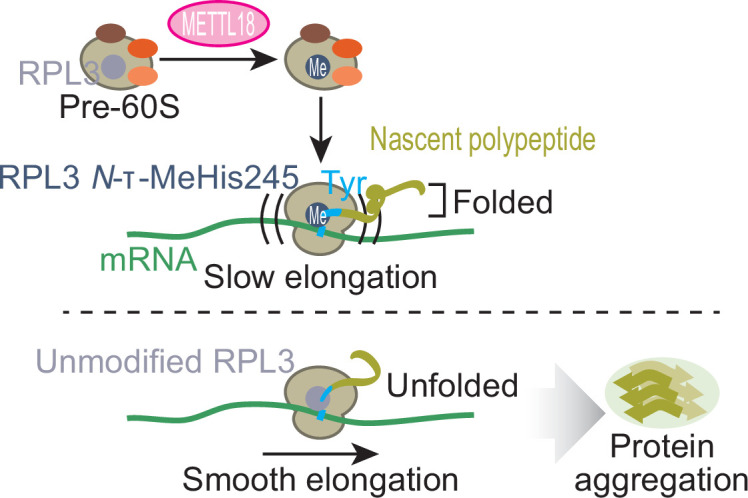
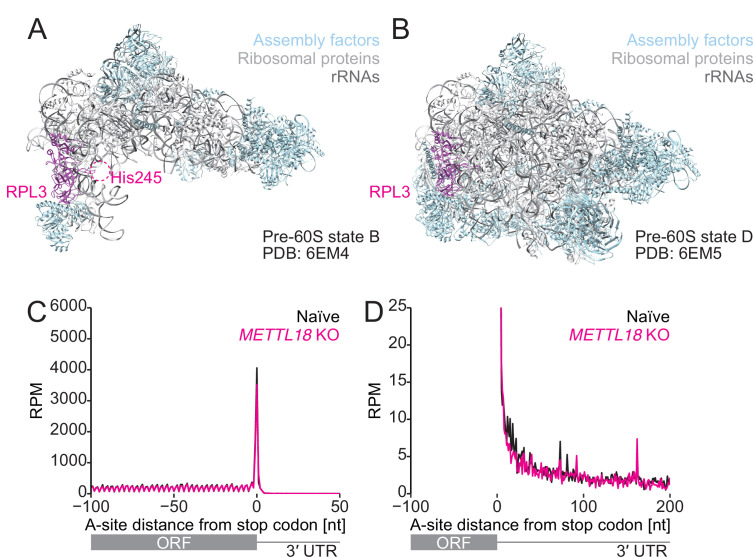
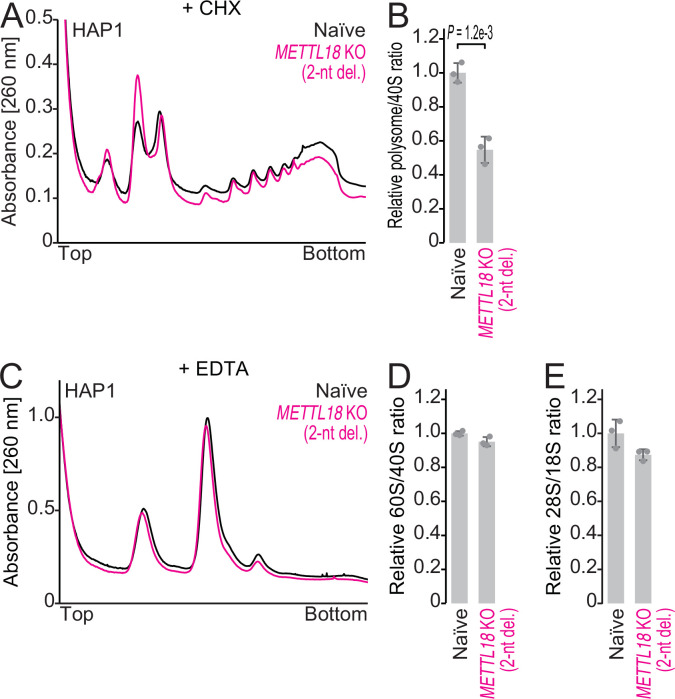
Figure 9. Schematic representation of METTL18-mediated control of translation and proteostasis.
METTL18 adds a methyl moiety at the τ-N position of His245 in RPL3 in the form of an early 60S biogenesis intermediate. Methylated ribosomes slow elongation at Tyr codons and extend the duration of nascent peptide folding, ensuring proteostatic integrity. Without RPL3 methylation, the accumulation of unfolded and ultimately aggregated proteins in cells was induced.