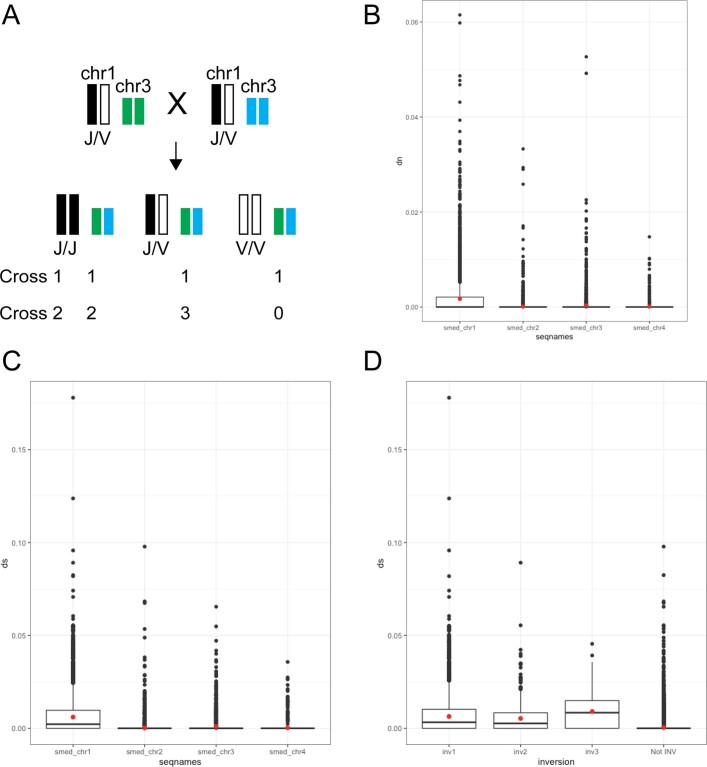
Extended Data Fig. 10. Embryonic lethality and synonymous divergence in Schmidtea mediterranea.
(A) Two J/V lines with differential SNP markers on chromosome 3 were crossed. Zygotes were collected for genotyping. Heterozygous chromosome 3 markers validate the occurrence of fertilization. All three genotypes of chromosome 1 (J/J, J/V and V/V) were observed in zygotes. (B-C) Non-synonymous divergence (B) and Synonymous divergence (C) for all heterozygous sites in the J/V strain S2 and their distribution in different chromosomes summarized as boxplots. Chromosome 1 has the highest mean (red dots) and median dN or dS values. The min, max, median, mean, first quartile, and third quartile of dN (B) for each chromosome are as following: chr1 = (0, 0.0052, 0, 0.0017, 0, 0.0021), chr2 = (0, 0, 0, 0.000092, 0, 0), chr3 = (0, 0, 0, 0.00031, 0, 0), and chr4 = (0, 0, 0, 0.000088, 0, 0). The min, max, median, mean, first quartile, and third quartile of dS (C) for each chromosome are as following: chr1 = (0, 0.024, 0.0022, 0.006, 0, 0.0098), chr2 = (0, 0, 0, 0.00021, 0, 0), chr3 = (0, 0, 0, 0.00093, 0, 0), and chr4 = (0, 0, 0, 0.00026, 0, 0). (D) Synonymous divergence for all heterozygous sites in the J/V strain S2, and their distribution in the three chromosome 1 inversions and the rest of the genome, summarized as boxplots. Mean (red dots) and median dS decreases in the following order: inversion 3 (inv3), inversion 1 (inv1), inversion 2 (inv2), and the rest of the genome (Not INV). One way ANOVA and Turkey Honest Significant Differences tests showed that dS were significantly different between all 3 inversions (inv2-inv1: p.adj = 0.005; inv3-inv1: p.adj < 1e-06; inv3-inv2: p.adj < 1e-06). The min, max, median, mean, first quartile, and third quartile of dS for each inversion are as following: inversion 1 = (0, 0.026, 0.0034, 0.0064, 0, 0.01), inversion 2 = (0, 0.021, 0.0026, 0.0053, 0, 0.0083), inversion 3 = (0, 0.036, 0.0084, 0.0091, 0, 0.015), and non-inversion = (0, 0, 0, 0.0005, 0, 0). A total of 8317 genes from chromosome 1, 6568 genes from chromosome 2, 3394 genes from chromosome 3, and 1834 genes from chromosome 4 were plotted for non-synonymous divergence (B) and synonymous divergence (C). A total of 6633 genes from inversion 1, 754 genes from inversion 2, 298 genes from inversion 3, and 12428 genes from the rest of the genome were plotted for synonymous divergence (D).