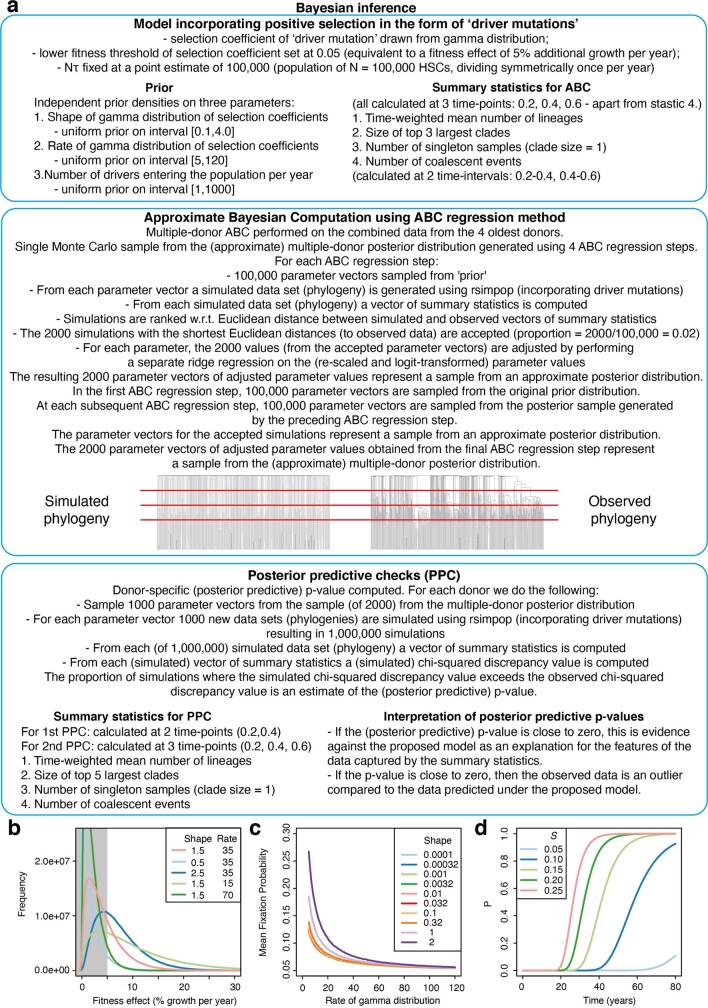
Extended Data Fig. 10. Modelling of HSC populations incorporating positive selection.
a, Overview of modelling approach used to estimate the shape and rate of the gamma distribution of selection coefficients from which ‘driver mutations’ are drawn, and the number of driver mutations drawn from this distribution (using a selection coefficient threshold of > 0.05) that are entering the HSC population per year. For these simulations Nτ was fixed at 100,000 and therefore only summary statistics for the first 3 timepoints were used to assess how well a given simulation for an individual resembled the observed tree. b, Plot showing maximum posterior density estimates of the rate and shape parameters of the gamma distribution for selection coefficients (pink line) obtained using Approximate Bayesian computation. Blue/green lines show how altering the rate and shape parameters affect the gamma distribution. c, Plot showing how changing the shape of the gamma distribution of selection coefficients (each line has a different shape) alters the probability of a driver gene fixing in the population. Reducing the shape below 0.1 does not affect the probability of driver gene fixation and therefore was the lower limit of the shape prior. d, Plot showing how the probability of detecting a clone with CF 2.5% changes over time for different selection coefficients. There is only a probability of 0.1 of being able to identify a driver mutation with a selection coefficient of 0.05 that entered the population at birth. We therefore used a lower threshold of 0.05 for the driver mutation selection coefficients.