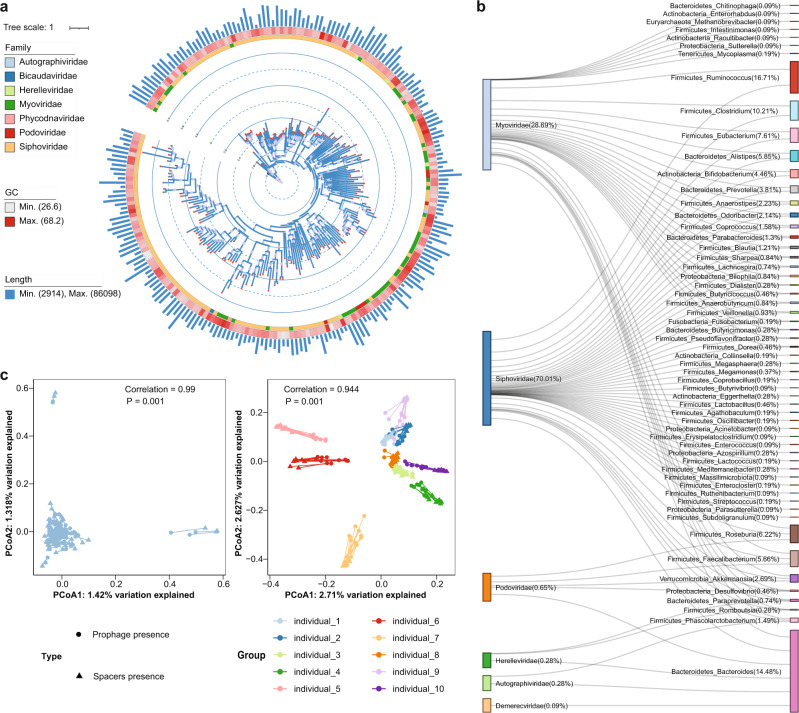
Fig. 4. ONT-improved metagenome contained highly diverse prophages and CRISPR spacers in human gut microbiome.
a Phylogenetic distribution of 228 prophages with both complete major capsid protein (MCP) and terminase large subunit (TLS) proteins (see Methods and Results section) discovered from 200 samples, with length distribution from 2.9 to 86 kb (scaled in barplot). For each sequence, the assigned viral family was indicated in color and the length of each prophage was shown as bar length in the outer circle. b Prophage-host pairs determined by analyzing prophage sequences and flanking regions. Prophages were grouped at the family level and bacteria at the genus level, with each side showing the percentage of family/genus among all the sequences. c Procrustes analysis of prophage/CRISPR spacer structures in the cross-sectional cohort (left) and time-series cohort (right), showing significant correlations between their overall compositions and indicating the highly stable prophage/CRISPR spacers structures within the same individual across time points. The statistical significance of the procrustes results were assessed using function protest with 999 permutations. The p value for both statistical tests was 0.001.