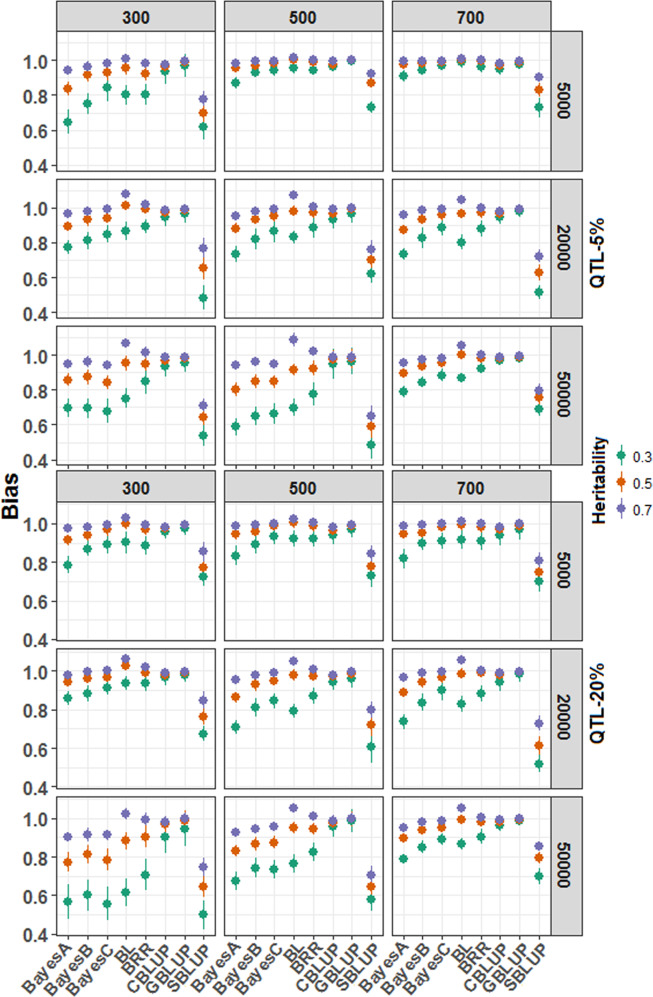
Fig. 6. The bias estimation in genomic estimated breeding value (GEBV) with the simulated dataset.
The bias is measured as the regression coefficients obtained by regressing the observed phenotypic values upon the predicted phenotypic values. Further, a repeated (100 times) fivefold cross-validation approach was adopted to compute the coefficients. The final coefficient estimates were computed by taking the average of the five folds and 100 replicates. The GBLUP is the least biased method for the GEBV prediction. GBLUP and CBLUP were identified as robust methods among the BLUP alphabets, irrespective of the trait genetic architecture. The BRR and BLASSO were less biased among the Bayesian models than the other variants.