Figure 1.
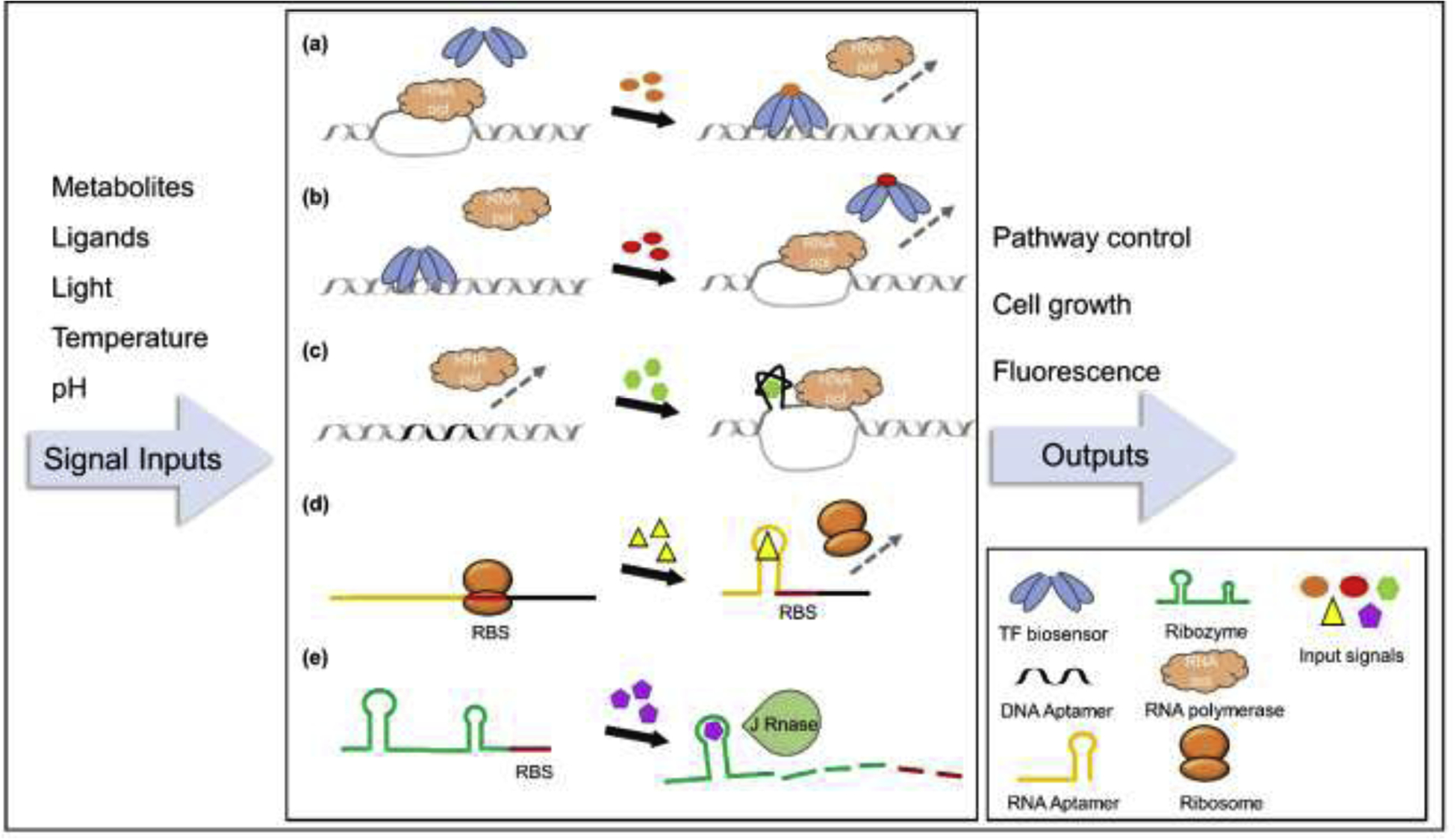
Application of TF and nucleic acid biosensors in pathway optimization and the regulation mechanisms. (a) TF biosensor-enabled transcription switch-on. The signal input activates the TF biosensor to interact with its corresponding promoter sequence, blocking the accession of RNA polymerase. (b) TF biosensor-enabled transcription switch-off. The signal causes some conformational changes of the TF biosensor, releasing it from the promoter and thus allowing the transcription by RNA polymerase. (c) DNA aptamer-promoted transcription switch-on. Signal input triggers the conformational changes of the DNA aptamer, promoting the unwinding of promoter sequence for RNA polymerase binding. (d) RNA aptamer-enabled translation switch-off. The conformational changes of the RNA aptamer inhibit the targeting of ribosome to RBS site. (e) Ribozyme-mediated translation switch-off. The ribozyme experiences self-cleavage with the signal input, leading to the digestion of the downstair mRNA. Input signals include diverse metabolites and environmental changes, enabling the generation of versatile regulation and screening strategies for metabolic engineering.
