Figure 2.
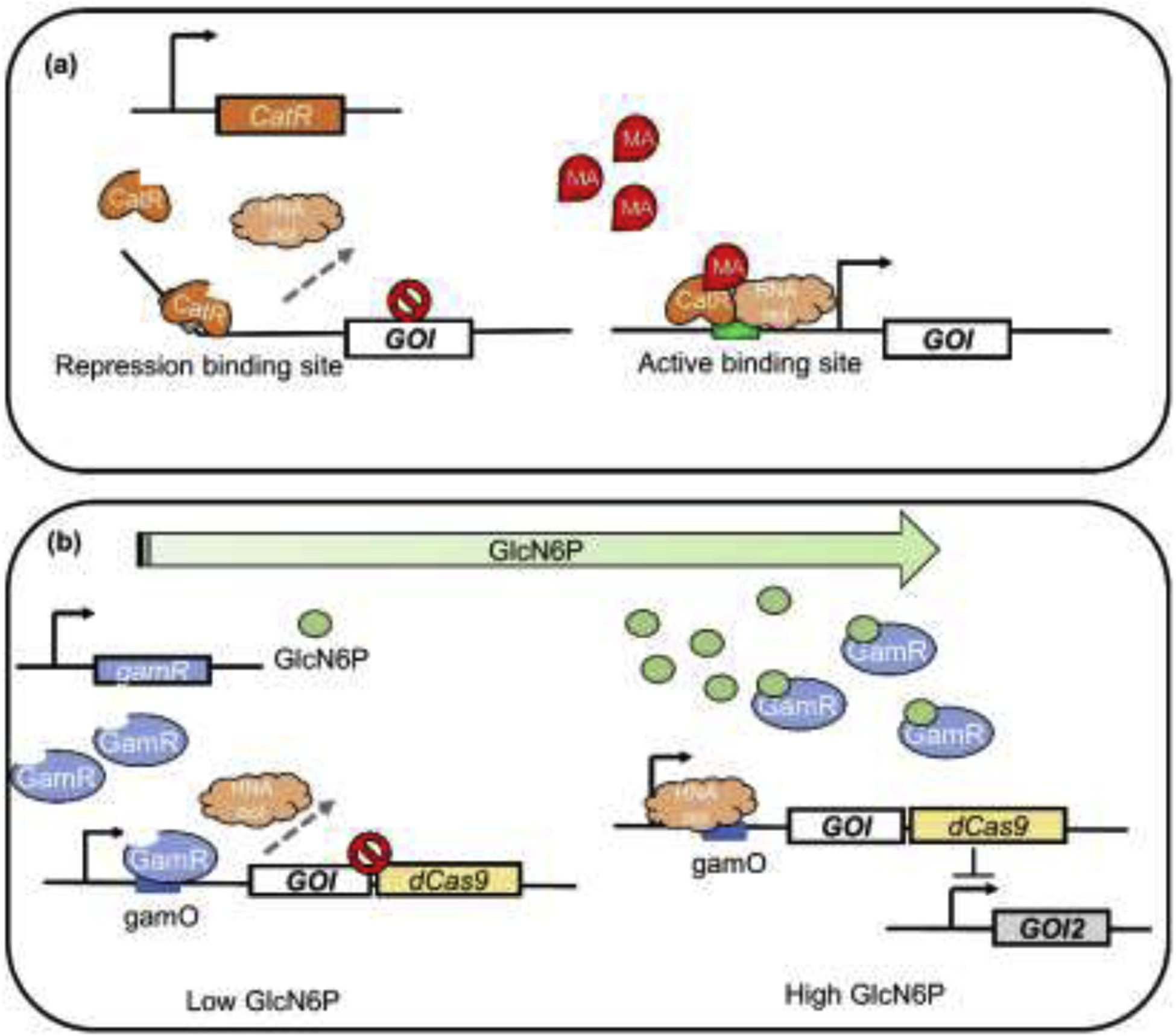
Application of metabolite-responsive biosensors for dynamic pathway regulation. (a) The mechanism of MA-responsive biosensor CatR. The CatR tetramer binds to a specific repression site, triggering the bending of DNA chain and blocking its interaction with RNA polymerase when the signal molecule MA is absent. Once MA combines with CatR and causes its conformational changes, CatR binds to another active site to release the bending of DNA and retrieve transcription. MA, muconic acid. (b) GlcN6P-responsive biosensor enabled bifunctional dynamic regulation. When the intracellular concentration of GlcN6P is low, TF biosensor GamR binds to gamO sites to block the binding of RNA polymerase, inhibiting the transcription of GOI (gene of interest) and CRISPR/dCas9. When GlcN6P concentration reaches high level, GamR combines with GlcN6P, and the allosteric effect of GlcN6P decreases the affinity of GamR to DNA and activates the expression of GOI and CRISPR/dCas9. The dCas9 coupled with sgRNA represses GOI2, thus realizing simultaneous activation and repression.
