Figure 1.
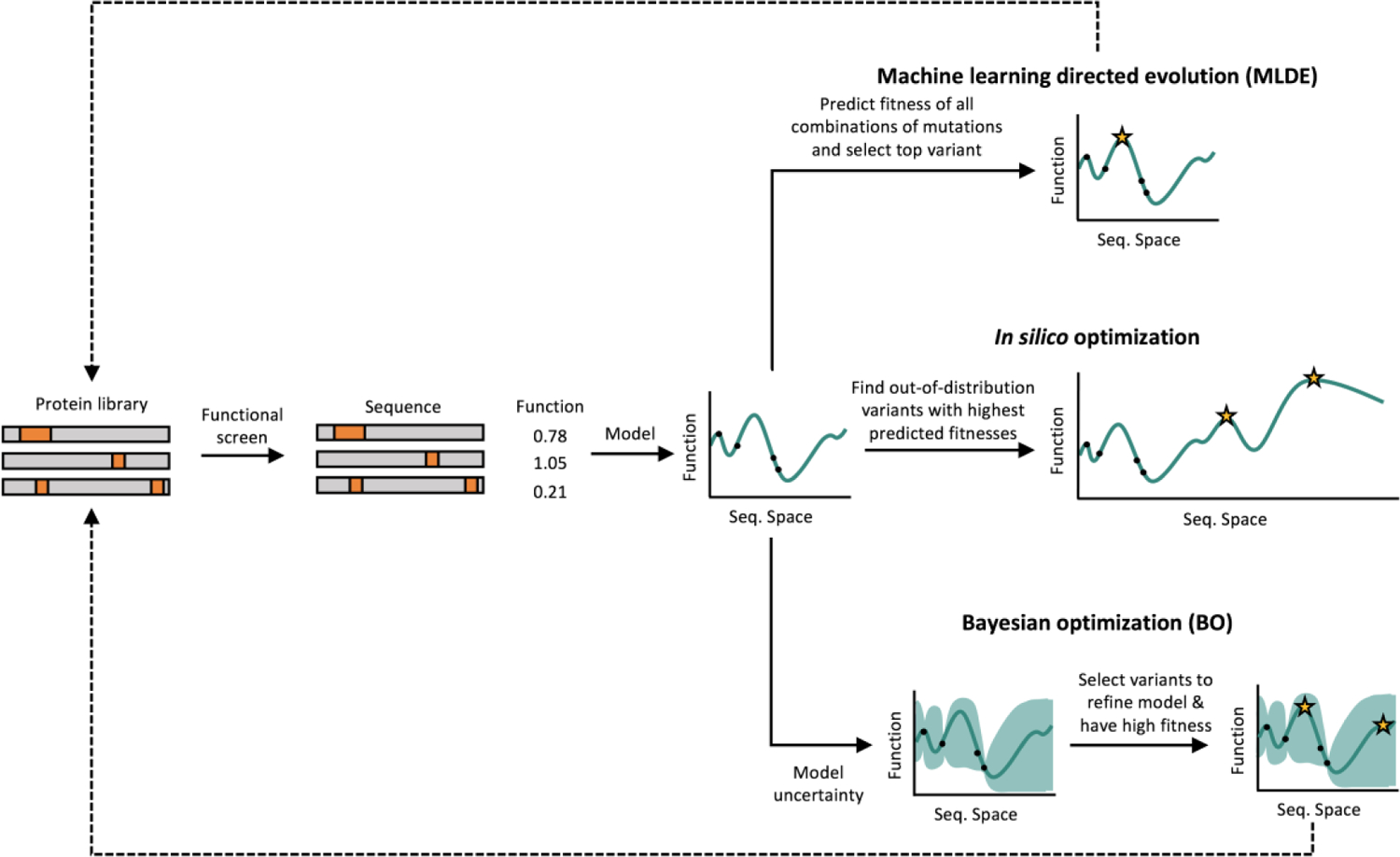
Machine learning-driven protein optimization strategies typically require an initial protein sequence library, typically created using error-prone PCR, site-saturated mutagenesis, or chimeragenesis. This protein sequence library is screened experimentally to determine a fitness value for each protein variant. Protein sequence-function data is modeled using supervised machine learning. The model can then be used to design improved variants in multiple ways. In machine learning directed evolution (MLDE), the fitness for other combinations of mutations in a combinatorial library are predicted and the best variant is selected for further rounds of mutagenesis, screening, and modeling. In silico optimization, however, uses optimization strategies to find highly fit protein variants far from the initial training library in the larger protein-sequence space. Bayesian optimization involves iterative rounds of mutagenesis, screening, and learning to maximize protein fitness over fewer rounds of experimental characterization by proposing protein variants that will help refine the model and have high fitness.
