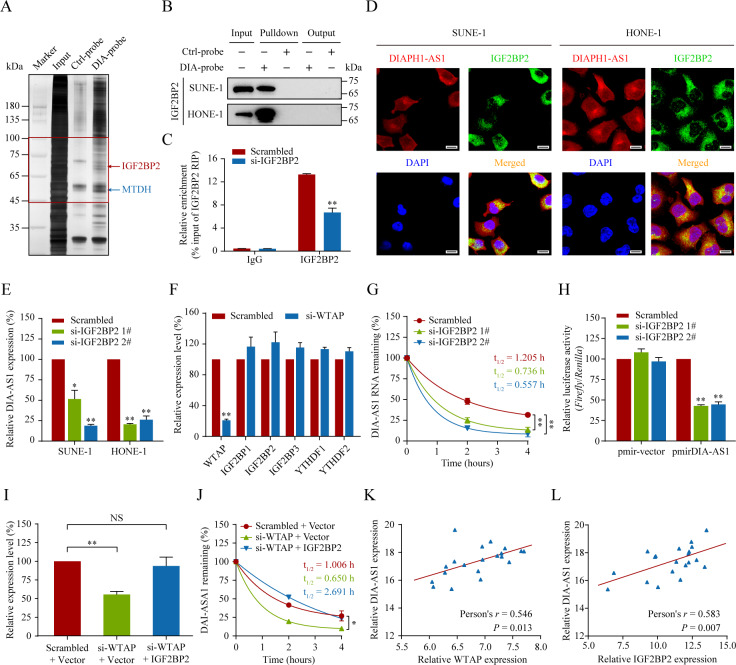
Fig. 5. IGF2BP2 interprets WTAP-mediated DIAPH1-AS1 m6A modification.
A, B Identification of the DIAPH1-AS1-protein complexes pulled down by DIAPH1-AS1 junction probes with proteins extracted from HONE-1 cells, followed by silver staining (A) and western blotting analysis (B). The specific bands (45–100 kDa, indicated in the red box) were cut off and subjected to mass spectrometry (MS) analysis. C IGF2BP2 was immunoprecipitated and RIP-qPCR was used to assess the association of DIAPH1-AS1 with IGF2BP2. D FISH and IF double staining in SUNE-1 and HONE-1 cells showing the co-localization of DIAPH1-AS1 (Cy3; Red) and IGF2BP2 (Green) in the cytoplasm; Nuclei are stained blue (DAPI). Scale bar: 20 μm. E Quantitative RT-PCR analysis of DIAPH1-AS1 expression in SUNE-1 and HONE-1 cells with or without WTAP silencing. F Quantitative RT-PCR analysis of mRNA levels of m6A readers (IGF2BP1, IGF2BP2, IGF2BP3, YTHDF1 and YTHDF2) in HONE-1 cells with or without WTAP silencing. G DIAPH1-AS1 RNA stability in control and IGF2BP2-silenced cells. Quantitative RT-PCR of DIAPH1-AS1 at the indicated time points after treatment with actinomycin D (10 μg/mL). H. Relative luciferase activity in HONE-1 cells co-transfected with luciferase reporter pmirGLO or pmirGLO-DIAPH1-AS1 and IGF2BP2 siRNAs or the control. I Quantitative RT-PCR analysis of DIAPH1-AS1 expression in HONE-1 cells co-transfected with si-WTAP and the IGF2BP2 overexpression vector or the corresponding control. J DIAPH1-AS1 RNA stability in HONE-1 cells co-transfected with si-WTAP and the IGF2BP2 overexpression vector or the corresponding control. Quantitative RT-PCR of DIAPH1-AS1 at the indicated time points after treatment with actinomycin D (10 μg/mL). K, L Pearson correlation analysis of DIAPH1-AS1 and WTAP mRNA (K), as well as DIAPH1-AS1 and IGF2BP2 mRNA (L) in 20 NPC tissues, determined using qRT-PCR. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01. The experiments were repeated at least three times independently.