Figure 10.
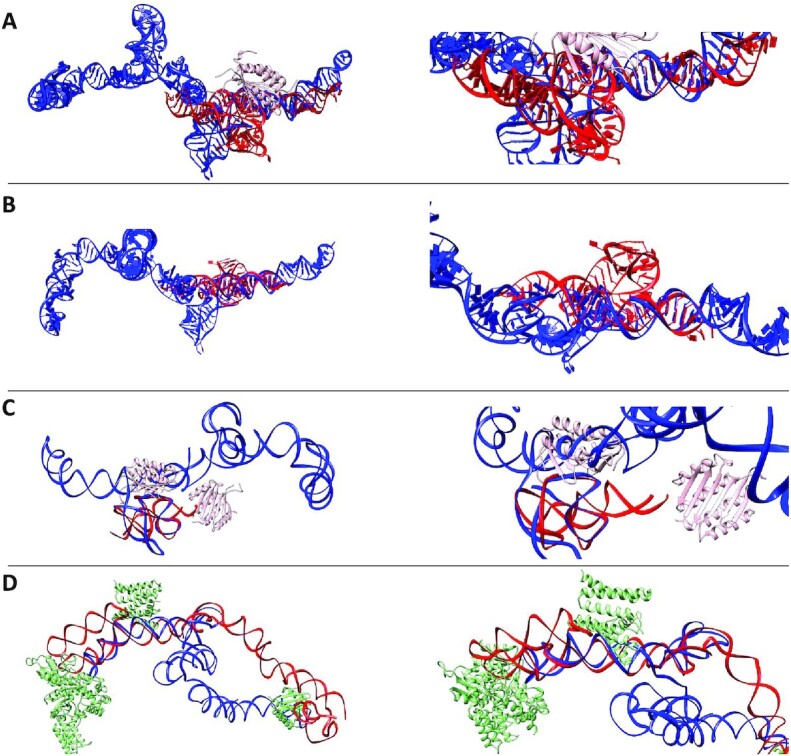
Superimposed Overlays of Alu Crystal Fragments with High-Resolution LincRNA-p21 Sense and Antisense Models. Left panels depict the overall structures and their overlays; right panels present the same but magnified view. (A) represents the crystal structure of archaeal (P. horikoshii) Alu RNA (Red, 4UYK) bound to the Signal Recognition Particle (SRP, 9 kDa) protein (Pink) and overlaid with the high-resolution 1036 model of sense LincRNAp-21 AluSx1 (Blue) (103) (see Supplementary Movie S7). (B) presents the crystal structure of Alu RNA derived from B. subtilis (Red, 4WFL), overlaid with the high-resolution 1036 model of sense LincRNA-p21 AluSx1 (Blue) (104) (see Supplementary Movie S8). (C) shows the overlap of the high-resolution LincRNA-p21 AluSx1 1036 model (Blue) with the high-resolution structure of human Alu RNA fragment (Red, 5AOX) bound to the SRP protein (Pink) (105) (see Supplementary Movie S9). (D) presents the high-resolution antisense LincRNA-p21 AluSx1 1013 model (Red) overlaid with the crystal structure of canine 7S RNA (Blue, 4UE5) bound to SRP protein (Green) (106) (see Supplementary Movie S10). Each Alu crystal fragment generally depicts strong alignment and conservation of secondary structure with the sense LincRNA-p21 AluSx1 model. RNAalign models were visualised by UCSF Chimera (107).
