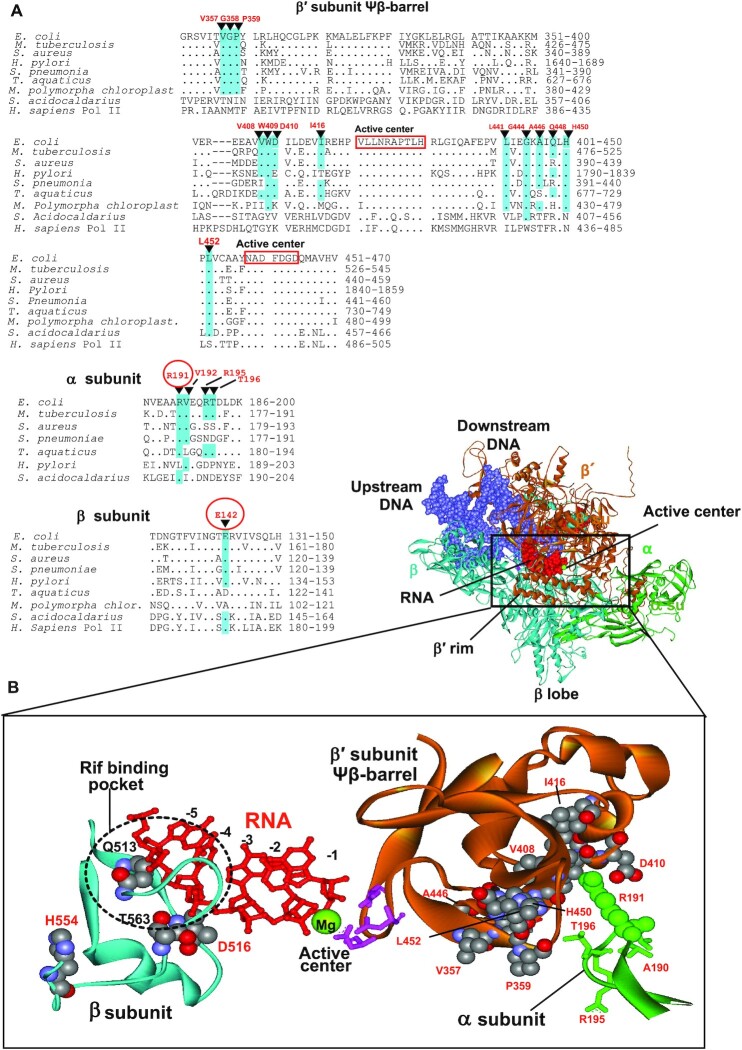
Figure 8.
Location of some Rifr compensatory mutations in Mtb (12) and E. coli (present work) (A) Sequence alignments of the regions of β′, β and α subunits of various organisms bearing compensatory mutations. Compensatory mutations are indicated. Mutations found in the present work are circled. Identical residues in the sequences are highlighted. (B) Structural context of the compensatory mutations. Location of the primary Rifr and compensatory mutations (both in space-fil representation) in the RNAP elongation complex is shown. Residues numbering is as in E. coli. Active center residues (pink) coordinating catalytic Mg2+ ion (green sphere) are indicated along with emerging RNA transcript (red). Position of Rif binding pocket is marked. Note, that all indicated residues are identical in E. coli and Mtb. E. coli RNAP structure is from (54).