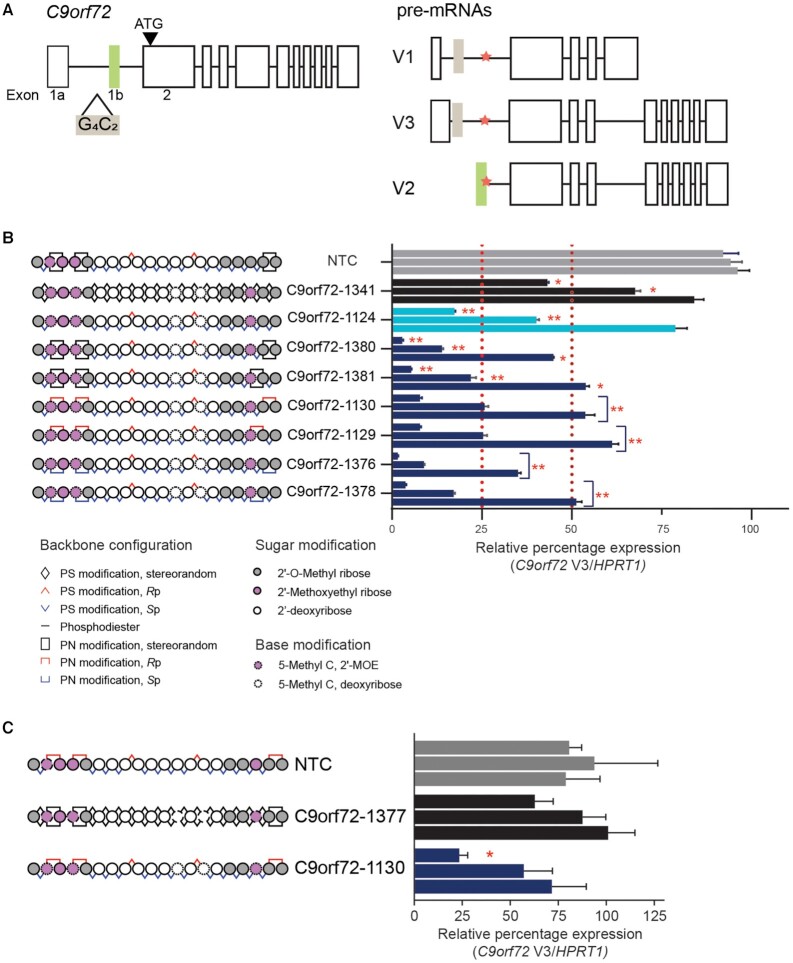
Figure 5.
Application of PN backbone chemistry improves potency of C9orf72-targeting oligonucleotides. (A) (Left) C9orf72 allele with hexanucleotide-repeat expansion mutation (G4C2, beige box) in intron 1 upstream of exon 1b (green box). (Right) Transcriptional pre-mRNA variants (V1, V2 and V3) produced by the mutated allele. V2 uses exon 1b as the first exon, whereas V1 and V3 use variations of exon 1a. Splice site 1b (SS1b, orange star) is at the exon 1b-intron 1 boundary. SS1b is accessible to antisense oligonucleotide in transcripts that use exon 1a (V1 and V3) but protected from degradation by the spliceosome in transcripts that use exon 1b (V2) (6). (B) Schematic representation of oligonucleotides (left) evaluated for activity against C9orf72 V3 in ALS motor neurons, including non-targeting control (NTC, 5′- CCTTCCCTGAAGGTTCCUCC-3′, gray), stereorandom control (C9orf72-1341, black), stereopure control (C9orf72-1124, light blue) and chimeric oligonucleotides with various combinations of PN linkages in the wings (navy blue). Oligonucleotides were delivered to ALS motor neurons under gymnotic conditions with concentrations increasing (0.08, 0.4, 2 μM) from bottom to top. Data are presented as mean ± sem, n = 3 per oligonucleotide per concentration. two-way RM ANOVA * P < 0.05, ** P< 0.01. (C) Schematic representation of oligonucleotides (left) evaluated for activity against C9orf72 V3 in ALS motor neurons, including non-targeting control (NTC, 5′-CCTTCCCTGAAGGTTCCTCC-3′, gray), fully stereorandom C9orf72-1377 (black) and fully stereopure C9orf72-1130 (navy). Oligonucleotides were delivered to ALS motor neurons under gymnotic conditions with concentrations increasing (0.08, 0.4, 2 μM) from bottom to top. Data are presented as mean ± sem, n = 3 per oligonucleotide per concentration.