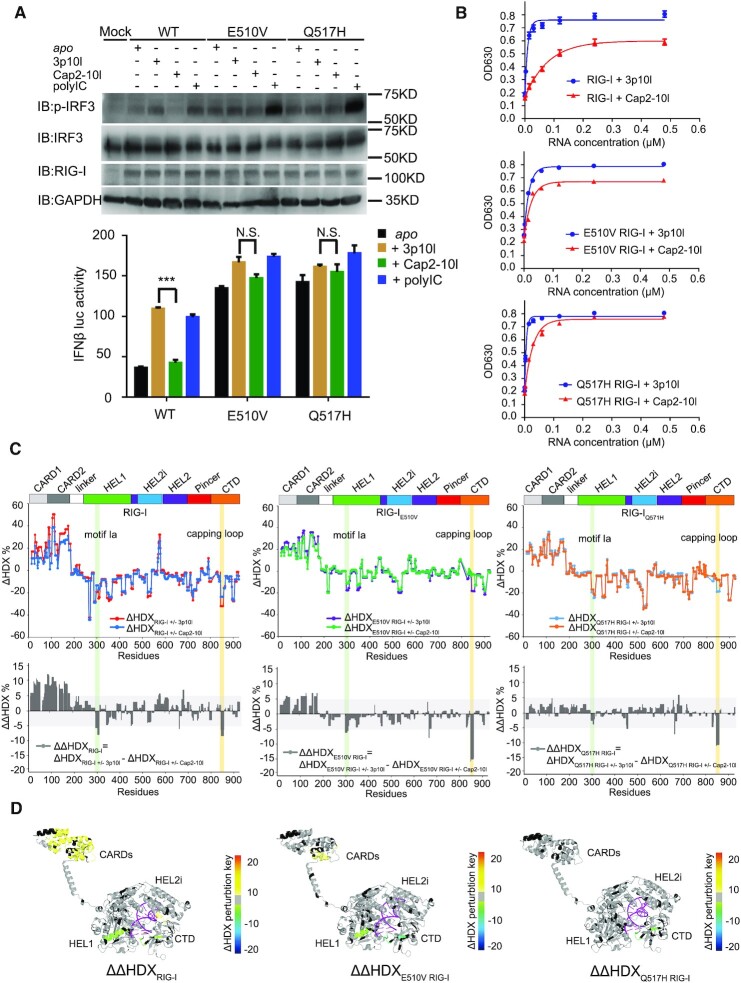
Figure 3.
Impaired RNA proofreading capabilities associated with RIG-IQ517H and RIG-IE510V. (A) Examination of IRF3 phosphorylation and IFN-β reporter assays of RIG-IE510V and RIG-IQ517H upon respective RNA treatment. Data are presented as the mean values ± SEM; n = 3 independent experiments; statistics indicate significance of differences between WT and the respective mutant determined by Student's t test: *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001. N.S. denotes nonsignificant. (B) RNA-dependent ATPase activity profiles of the indicated protein complexes. (C) Single amide consolidated HDX-MS profiling of RIG-I, RIG-IE510V and RIG-IQ517H upon RNA discrimination between 3p10l and Cap2-10l. In the uppor panel, the y- and x-axes illustrate the HDX perturbations (ΔHDX%) of respective RNA binding event and domain arrangement of RIG-I or its HEL2i variants, respectively. A positive value or a negative value in the y axis (ΔHDX%) represents protection or deprotection against deuterium exchange in the corresponding region of the receptor depicted in the x axis when a RNA binding event (induced by 3p10l or Cap2-10l) takes place. In the lower panel, it displays the comparative HDX analysis (ΔΔHDX%) between 3p10l- and Cap2-10l-mediated HDX perturbations on RIG-I, RIG-IE510V and RIG-IQ517H. ΔΔHDX% values (ΔΔHDX = ΔHDXRIG-I(WT/mut) ± 3p10l - ΔHDXRIG-I(WT/mut) ± Cap2-10l) could indicate the conformational perturbations of RIG-I or its variants upon RNA discrimination. (D) Differential single amino acid consolidation HDX data are mapped onto the full-length RNA-bound RIG-I structure model in the ribbon, as shown by the representation of ΔΔHDX profiles of RIG-I, RIG-IE510V and RIG-IQ517H (Supplementary Figure S5, columns (xiii, xiv and xv), and Figure S8). Percentages of deuterium differences are color-coded according to the HDX key (below). Black, regions that have no sequence coverage and include proline residues that have no amide hydrogen exchange activity; gray, no statistically significant changes between compared states; purple, duplex RNA ligand.