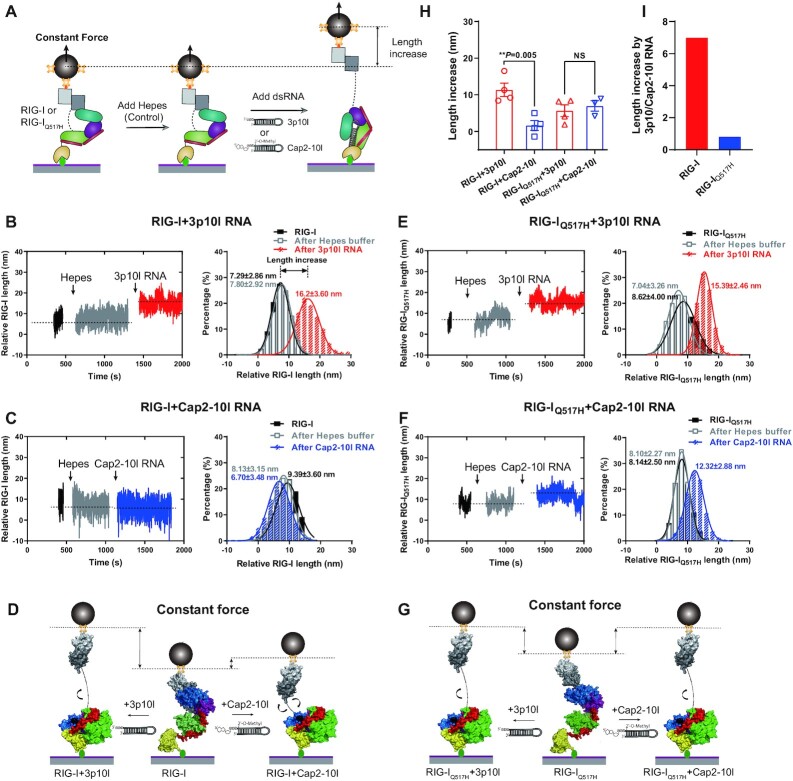
Figure 4.
RIG-IQ517H extends its conformation unbiasedly upon RNA proofreading. (A) Schematic revealing the RIG-I and RIG-IQ517H discrimination of viral and self RNAs in MT. When RIG-I and RIG-IQ517H were captured with a low force (∼1.3 pN) in MT, HEPES buffer flowed in as the control, and then the length increase of RIG-I or RIG-IQ517H driven by RNAs was measured. (B, C) Representative data of the length increase of RIG-I driven by 3p10l RNA (B) and RIG-I driven by Cap2-10l RNA (C), wherein the relative RIG-I length under different conditions was quantified by Gaussian fitting. (D) Schematic of the length increase of RIG-I driven by 3p10l RNA or Cap2-10l RNA. (E, F) Representative data of the length increase of RIG-IQ517H driven by 3p10l RNA (E) and RIG-IQ517H driven by Cap2-10l RNA (F), wherein the relative RIG-IQ517H length under different conditions was quantified by Gaussian fitting. (G) Schematic of the length increase of RIG-IQ517H driven by 3p10l RNA or Cap2-10l RNA. (H) Length increases in RIG-I and RIG-IQ517H driven by 3p10l RNA or Cap2-10l RNA. Each dot is derived from repeated experiments. (I) Length increases driven by 3p10l RNA and Cap2-10l RNA. Data are presented as the mean ± SD in (B), (C), (E) and (F) and as the mean ± SEM in (H); statistics indicate significance of differences between compared sample states determined by Student's t test: **P ≤ 0.01; NS represents nonsignificant in (H).