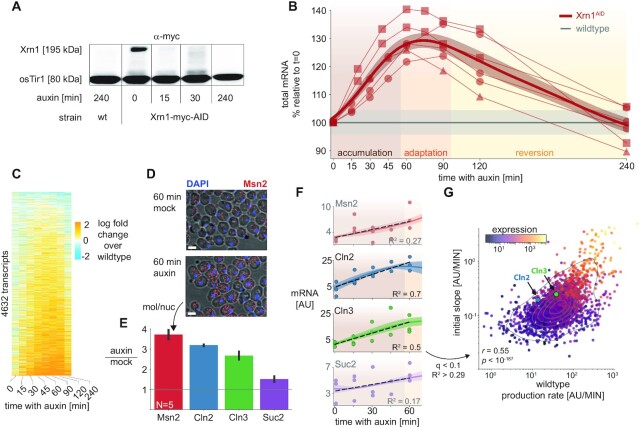
Figure 3.
Xrn1 acute depletion causes a transient increase in mRNA levels. (A) Auxin inducible degradation (AID) of Xrn1 is rapid and stable. Western blot (anti-myc) for Xrn1 tagged with an auxin-inducible degron (AID) and a myc-tag. Shown are the isogenic untagged strain (left), and a time course that demonstrates virtually no Xrn1 protein within 15 min. osTir1 is also tagged with Myc in these strains and is used as a loading control. See also Supplementary Figure S5G. (B) Accumulation of mRNA immediately following Xrn1 depletion. mRNA counts (scaled by spike-in reads) from the Xrn1 depletion time course experiment (x-axis). mRNA is scaled to initial values, and to the corresponding wildtype measurement (y-axis) in a total of six biological replicates (lines) in three different experimental batches (markers). The gray line indicates the wildtype trajectory under the same transformation with standard deviation as a shaded area. The thick red line represents the average of the smoothed interpolations of each separate trajectory. We label the three stages of the response for convenience (accumulation → adaptation → reversion). (C) Genome-wide mRNA accumulation and reversion. mRNA per transcript (y-axis) was normalized with the corresponding measurement from the wildtype time course (time along the x-axis), and the log fold change is color-coded. Transcripts were filtered to not have any missing values along the trajectory (N = 4632, ∼70%), and were sorted by their average log fold change between 15 and 120 min. (D) Single-molecule FISH validation. Composite micrographs (from a confocal Z-stack image), showing DIC image of cells with a max-projection of DAPI stain in blue and fluorescent probes for Msn2 in red (1 μm scale bar). Individual molecules and nuclei are discernible and are counted. Xrn1AID cells were treated with auxin or mock (DMSO) for 60 minutes, fixed, stained, and imaged. A clear increase in molecule counts is observed. (E) Single-molecule FISH quantification. In each field (N = 5), the number of observed molecules is divided by the number of observed nuclei to estimate the mRNA content of each cell in four different probes (x-axis). The y-axis denotes the ratio of the mRNA content in auxin versus mock treatment. (F) scaled RNA trajectories, and rate of accumulation. Each subplot shows the (spike-in scaled) mRNA counts (y-axis, points) from three replicates in the same experiment along the time course following Xrn1 depletion (x-axis). The line is the mean (+SEM) over interpolations of each separate repeat (N = 3). Selected transcripts correspond to smFISH probes (D, E). The dashed black line indicates the linear fit for the accumulation phase of the response (R2 noted per gene). Only fits within the 10% FDR threshold (R2 > 0.29, see S3E) are plotted in (G), Msn2 and Suc2 are shaded and do not appear in (G) as they are below this threshold. (G) mRNA accumulation correlates to transcript production rate. Comparing slopes from the FDR-selected linear fits (y-axis), to the production rate estimated from the wildtype sample (Figure 2D), 30% of the observed variability (R2) can be attributed to the production rate (P < 10–187). Note that there is a strong correlation between the two measures and the overall expression (color-coded, log scale), see text and Supplementary Figure S3F, G.