Figure 5.
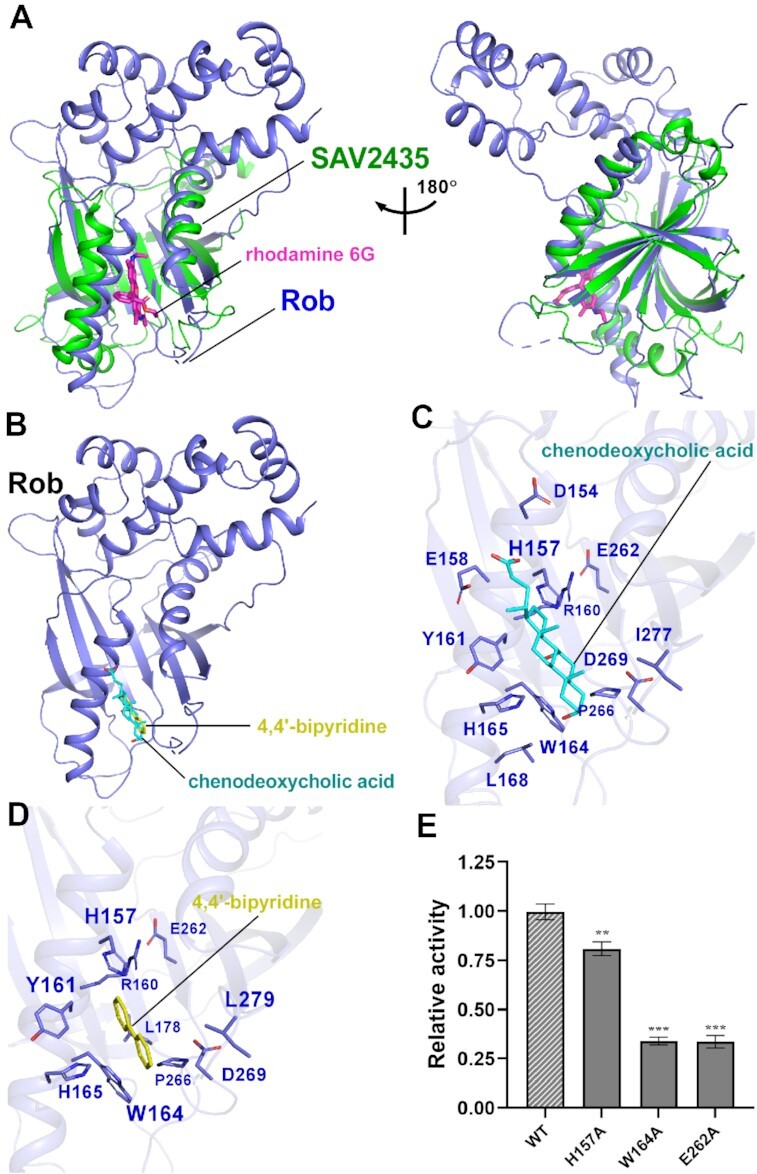
Molecular docking analysis of Rob with potential ligands. (A) Structural superimposition of the Gyrl-like family protein SAV2435 (PDB ID: 5KAW) onto the Rob from Rob-TAC. Rob and the SAV2435 are shown as blue and green cartoon. The ligand rhodamine 6G(R6G) is shown in magenta stick. (B) Rob docked with chenodeoxycholic acid and 4,4′-bipyridine. (C) Predicted binding pockets for chenodeoxycholic acid on Rob protein. Residues potentially interacted with chenodeoxycholic acid are shown in blue sticks. (D) Predicted binding pockets for 4,4’-bipyridine on Rob protein. Residues potentially interacted with 4,4’-bipyridine are shown in blue sticks. (E) Substitutions of conserved residues involved in Rob-ligand interactions suppressed in vitro transcription activity in the presence of 100 μM chenodeoxycholic acid. Data for in vitro transcription assays are means of three technical replicates. Error bars represent ± SEM of n = 3 experiments. Asterisk (***) indicates highly significant (P value < 0.001) difference from the wild-type Rob analyzed by one-way ANOVA with Tukey's multiple comparison test, respectively.
