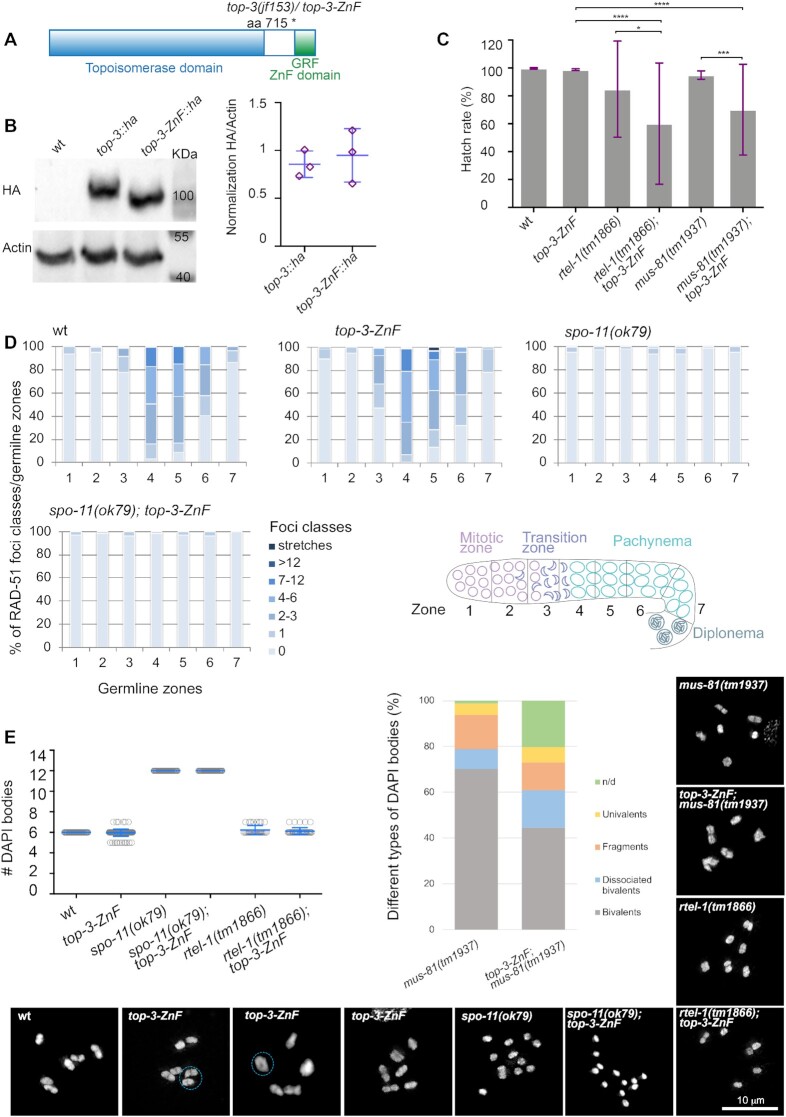
Figure 1.
t op-3-ZnF is a hypomorph allele. (A) Schematic representation of the top-3 gene, depicting the conserved topoisomerase domain (light blue) and the GRF zinc finger (ZnF) motif (green). The jf153 (top-3-ZnF) allele encodes a truncated protein that lacks the ZnF motif. (B) Western blot analysis of TOP-3::HA and TOP-3-ZnF::HA in whole worm extracts. wild-type (wt) worms were used to control for the specificity of the antibody. The TOP-3::HA protein is 100 KDa in size and the TOP-3-ZnF::HA protein lacks 45 amino acids. Actin was used as the loading control. Right panel: western blot quantification of three biological replicates per sample. Error bars indicate the mean and SD. (C) Hatch rates for the indicated genotypes: wt, 99.8% ± 0.3%, n = 18; top-3-ZnF, 98.8% ± 0.7%, n = 17; rtel-1(tm1866), 84.7% ± 34.3%, n = 8; rtel-1(tm1866); top-3-ZnF, 60.1% ± 43.4%, n = 9; mus-81(tm1937), 95.0% ± 3.0%, n = 8; mus-81(tm1937); top-3-ZnF, 70.1% ± 32.5%, n = 7. n = number of hermaphrodites. The histogram indicates the mean ± (SD). * P = 0.02, *** P = 0.0003, **** P < 0.0001, as determined using the Mann–Whitney test; non-significant differences are not shown. (D) The percentage of RAD-51 foci classes in each of the seven zones of the gonad in the indicated genotypes. For each zone, the average number of RAD-51 foci/nucleus was calculated from three gonads per genotype. Bottom right panel: schematic representation of the C. elegans gonad divided into seven equal zones. (E) Quantification of DAPI bodies in the −1 diakinesis oocyte in the indicated genotypes. Mean ± SD and number (n) of DAPI bodies per genotype: wt, 6 ± 0, n = 50; top-3-ZnF, 5.96 ± 0.3, n = 154; spo-11(ok79), 12 ± 0, n = 50; spo-11(ok79); top-3-ZnF, 12 ± 0, n = 50; rtel-1(tm1866), 6.2 ± 0.4, n = 34; and rtel-1(tm1866); top-3-ZnF, 6.1 ± 0.3, n = 41. Right panel: qualitative analysis of the −1 diakinesis oocyte of the indicated genotypes. n = number of DAPI bodies and the percentage of the different types of DAPI bodies observed per genotype: mus-81(tm1937), n = 90, bivalents = 70.2%, dissociated bivalents = 8.5%, DNA fragments = 14.9%, univalents = 5.3%, n/d (not defined) = 1.1%; mus-81(tm1937); top-3-ZnF, n = 50, bivalents = 44.6%, dissociated bivalents = 16.2%, fragments = 12.2%, univalents = 6.8%, n/d (not defined) = 20.3%. Insets show representative images of DAPI-stained diakinesis nuclei of the indicated genotypes. For top-3-ZnF, the dashed light blue circles indicate the presence of bridges and a doughnut-shaped bivalent.