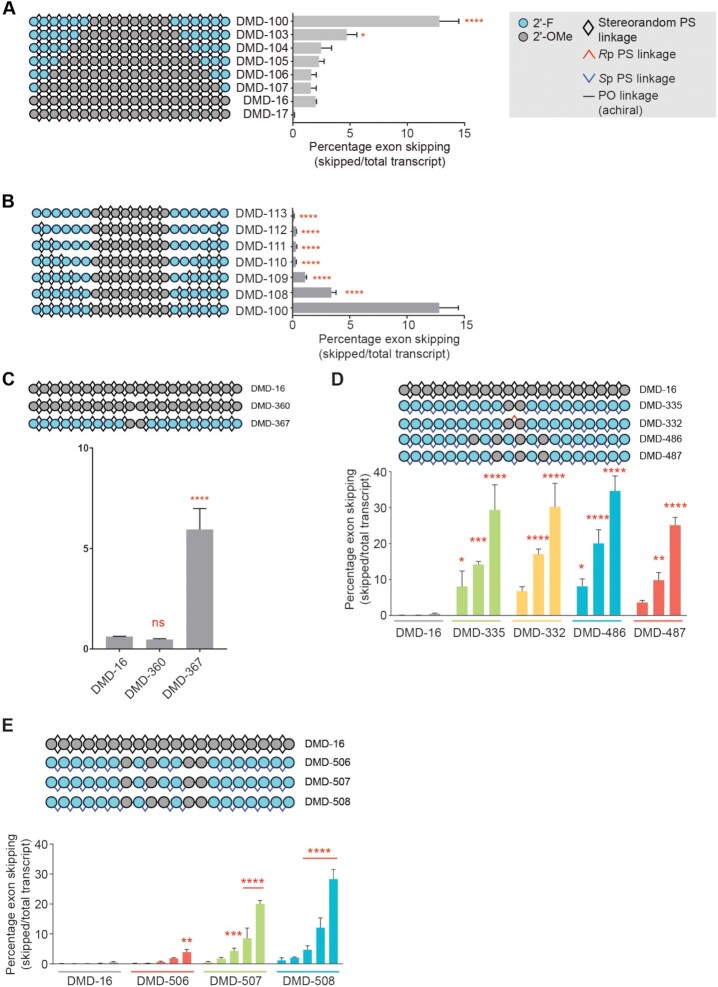
Figure 2.
Characterization and optimization of exon skipping oligonucleotide chemistry for human DMD exon 51. Quantification of exon 51 skipping in human Δ48–50 myoblasts treated gymnotically with the indicated oligonucleotide (10 μM A-C; dose response 0.1, 0.3, 1, 3 or 10 μM D,E). (A) Comparison of activities for oligonucleotides with identical sequence and backbone stereochemistry (stereorandom PS at all positions) with variable numbers of 2′-F modifications in termini. DMD-17 is complementary to mouse Dmd exon 23 and serves as non-targeting control in this experiment. (B) Evaluation of the effect of PO linkages on activity of oligonucleotides with identical sequence and 2′ ribose modification pattern **** P < 0.0001, * P < 0.05 One-way ANOVA with post-hoc comparison to DMD-16 (panel A) or to DMD-100 (panel B), Mean ± s.e.m., n = 2. (C) Evaluation of relationship between PO linkages and 2′-ribose modifications in the center of the oligonucleotide and its activity. **** P < 0.0001, ns, non-significant, One-way ANOVA with post-hoc comparison to DMD-16, Mean ± s.e.m., n = 2. (D) Application of Sp PS backbone stereochemistry and exploration of various 2′-ribose modification patterns in the center of the oligonucleotide. **** P < 0.0001, *** P < 0.001, **P < 0.01,* P < 0.05 Two-way ANOVA with post-hoc comparison to DMD-16, Mean ± s.e.m., n = 2. (E) Impact of PO linkages at various backbone positions on activity. **** P < 0.0001, *** P < 0.001, **P < 0.01,* P < 0.05 Two-way ANOVA with post-hoc comparison to DMD-16, Mean ± s.e.m., n = 2.