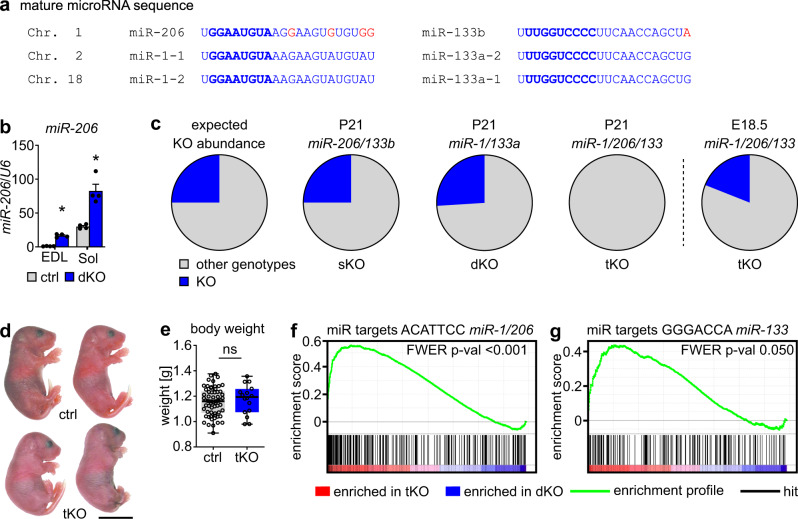
Fig. 1. miR-206/133b compensate for the absence of miR-1/133a in skeletal muscles to prevent perinatal death.
a The miRNAs miR-1, -133a/b and -206 are encoded in three clusters located on mouse chromosome (Chr.) 1, 2 and 18. The primary sequence of mature miR-1/133a is identical; miR-206 varies from miR-1 only by four bases, miR-133b differs in only one base from miR-133a (red). The seed sequence, essential for miRNA-target interaction, is identical in miR-1/206 and miR-133a/133b, respectively (bold blue). b Relative expression of miR-206 in wild-type (control) and adult miR-1/133a dKO extensor digitorum longus muscles (EDL) and soleus muscles (Sol) analyzed via TaqMan assay (n = 4 ctrl/4 dKO, n = 4 ctrl/4 dKO [males/females, 13–17 weeks], Mann-Whitney-U test, one-tailed, *p = 0.0143, Data are presented as mean values +/− SEM). U6 snRNA served as control. c Percentages of miR-206/133b sKO and miR-1/133a dKO animals at postnatal day 21 (P21) are in accordance to the expected Mendelian distribution (likelihood 25%), while miR-1/206/133 tKO animals were missing at P21. Analysis of E18.5 animals reveals presence of miR-1/206/133 tKO animals at this developmental stage (P21: n = 147 control littermates 48 sKO, n = 100 control littermates/35 dKO, n = 34 control littermates/0 tKO at P21; E18.5: n = 26 control littermates 6 tKO). d Gross morphology of control and miR-1/206/133 tKO pups after caesarean section (control = littermates, scale-bar: 1 cm [E18.5]). e Box-plot of miR-1/206/133 tKO and control body weight (n = 63 control/16 tKO, control = littermates, Mann-Whitney-U test, one-tailed, ns = not significant, Box indicates median and 25th to 75th percentiles, whiskers indicate minimum and maximum [E18.5]). f, g GSEA of miR-1/133/206 tKO compared to miR-1/133a dKO quadriceps muscle transcriptome data reveals enrichment of predicted miR-1/206 and miR-133 targets in miR-1/133/206 tKO (n = 4 dKO/4 tKO, familywise-error rate method (FWER), p = <0.001 (f), p = 0.05 (g) [E18.5]). Source data are provided as a Source Data file.