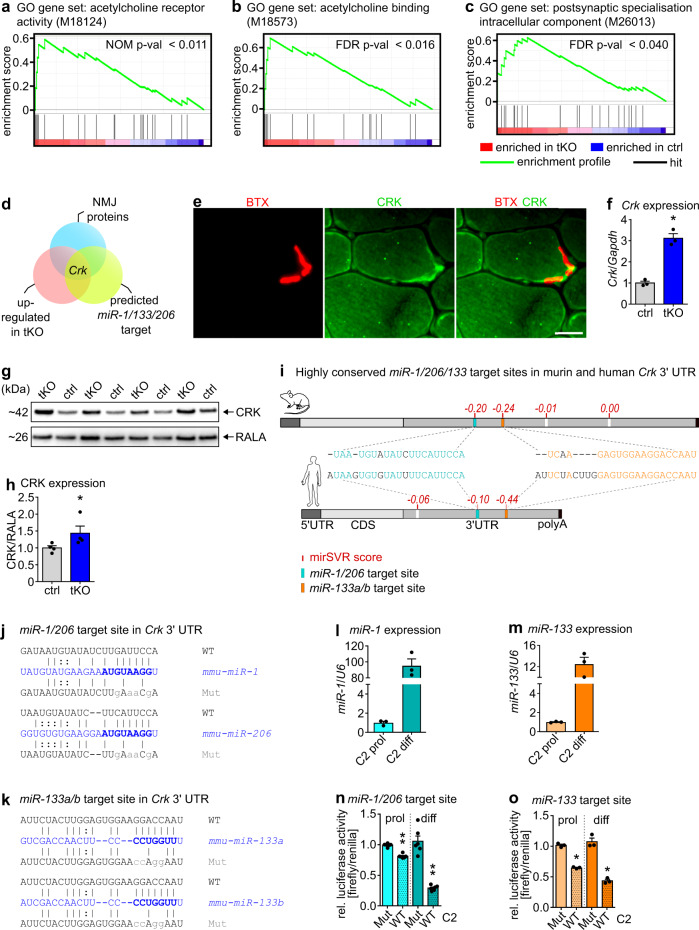
Fig. 3. The NMJ associated adapter protein Crk contains evolutionary conserved miR-target sites in the 3´UTR and is a target for miR-1/206/133.
a–c GSEA of control and miR-1/206/133 tKO quadriceps muscle transcriptome data (n = 4 control/4 tKO, control = miR-1-1/133a-2+/+//miR-1-2/133a-1lox/lox//miR-206/133b+/+//Pax7-Cre+/+, familywise-error-rate method (FWER), p = <0.011 (a), p = <0.016 (b), p = <0.040 (c) [E18.5]). GO = Gene Ontology. d Overlap of transcriptome data (significantly up-regulated in tKO with fold change >1.35) and predicted miR-1/206/133 target genes identified the NMJ-associated gene Crk as a top candidate. e CRK immunostaining reveals localization at the postsynapse in transversal sections of WT TA-muscles (scale-bar: 20 µm, BTX = bungarotoxin, n = 2 [male, 14 weeks]). f Crk expression in miR-1/206/133 tKO deficient muscles compared to control quadriceps (TaqMan assay, n = 3 control/3 tKO, control= miR-1-1/133a-2+/+//miR-1-2/133a-1lox/lox//miR-206/133b+/+//Pax7 Cre+/+, Mann-Whitney-U test, one-tailed, *p = 0.05, [E18.5]). Gapdh served as control. g, h Western blot analysis of CRK expression in miR-1/206/133 deficient diaphragm muscles (tKO) compared to control. RALA served as control. (n = 4 control/4 tKO, control= miR-1-1/133a-2+/+//miR-1-2/133a-1lox/lox//miR-206/133b+/+//Pax7-Cre+/+, Mann-Whitney-U test, one-tailed, *p = 0.0143, [E18.5]). i Conserved predicted binding sites in murine and human Crk mRNA. Putative miR-1/206 target sites (turquoise) and miR-133 binding sites (orange) in the 3´UTR. MicroRNA target sites ranked by mirSVR down-regulation score (red). CDS = coding sequence, UTR = untranslated region, polyA= polyadenylation-tail. j, k WT and mutated (Mut) sequences (gray) of putative miR-1/206 (j) and miR-133a/b (k) target sites in the 3´UTR of Crk were cloned into luciferase reporter vectors. MiR-sequence (blue), seed-sequence (bold blue). l, m Endogenous miR-1 or miR-133 expression in proliferating (prol) and differentiation stages (diff) of C2 cells (TaqMan assays, n = 3 prol/3 diff, n = 3 prol/3 diff, biological replicates). U6 served as control. n, o Repression of firefly-luciferase activity in Dual-Luciferase reporter assays by WT but not mutated (Mut) miR-1 or miR-133 miRNA target sites from 3´UTR of Crk. Repression of firefly-luciferase by the respective WT target sites is detected in proliferating, but is more pronounced in differentiating (diff) C2 cells (miR-1/206 target site n = 6 Mut/6 WT, miR-133 target site n = 3 Mut/3 WT, biological replicate = independent transfection, Mann-Whitney-U test, one-tailed, **p = 0.0011, *p = 0.05). Normalization via renilla-luciferase (endogenous control). Source data are provided as a Source Data file. Data are presented as mean values +/−SEM in f–h and l–o.