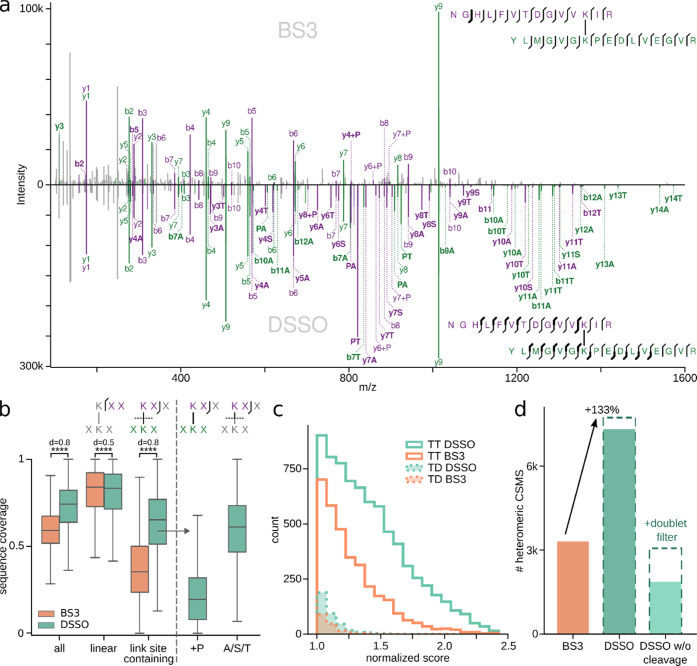
Figure 4.
Comparison of noncleavable crosslinker BS3 to the MS-cleavable crosslinker DSSO. (a) Example MS2 spectrum of a high-scoring CSM identified in both datasets. (Top) CSM from the BS3 dataset. (Bottom) The same peptide m/z-species identified in the DSSO dataset. Unique fragments are highlighted in bold. (b) Sequence coverage of all, linear and link site-containing fragments of all common CSMs (n = 12919). For DSSO, link site-containing fragments are additionally separated into fragments containing the full second peptide (+P) or only the cleaved crosslinker stub (A/S/T). Boxplots depict the median (middle line), upper and lower quartiles (boxes), and 1.5 times the interquartile range (whiskers). Asterisks indicate significance calculated by a two-sided Wilcoxon signed rank (p-value < 0.0001: ****). (c) Target–target and target–decoy score distributions of heteromeric CSMs for BS3 and DSSO. Scores were normalized to their respective score cutoff at 10% FDR. (d) Number of heteromeric CSMs passing 5% CSM-level FDR for BS3 and DSSO. As a control, DSSO was additionally searched as a noncleavable crosslinker and also filtered for the presence of peptide doublets.