Abstract
Enterobacter cloacae A-11 is a prototrophic, glycolytic mutant of strain 501R3 with a single transposon insertion in pfkA. The populations of strain A-11 on cucumber and radish seeds were smaller than the populations of strain 501R3 in natural soil, but the populations of these two strains on pea, soybean, sunflower, and sweet corn seeds were similar (D. P. Roberts, P. D. Dery, I. Yucel, J. Buyer, M. A. Holtman, and D. Y. Kobayashi, Appl. Environ. Microbiol. 65:2513–2519, 1999). The net effect of the mutation in pfkA in vitro was a shift from rapid growth on certain carbohydrates detected in seed exudates to much slower growth on other carbohydrates, amino acids, and organic acids. The impact of the mutation in pfkA was greatest on the growth rate of E. cloacae on the seeds that released the smallest quantities of fructose, other carbohydrates, and amino acids. Corn, pea, soybean, and sunflower seeds released total amounts of carbohydrates and amino acids at rates that were approximately 10- to 100-fold greater than the rates observed with cucumber and radish seeds for the first 24 h after inhibition began. The growth rate of strain A-11 was significantly less (50% less) than the growth rate of strain 501R3 on radish seeds, and the growth rate of strain A-11 was too low to estimate on cucumber seeds in sterile sand for the first 24 h after inhibition began. The growth rate of strain A-11 was also significantly lower on soybean seeds, but it was only 17% lower than the growth rate of strain 501R3. The growth rates of strains 501R3 and A-11 were similar on pea, sunflower, and corn seeds in sterile sand for the first 30 h after imbibition began. Large reductions in the growth rates of strain A-11 on seeds were correlated with subsequent decreased levels of colonization of seeds compared to the levels of colonization of strain 501R3. The strain A-11 populations were significantly smaller than the strain 501R3 populations only on radish and cucumber seeds. The mutation in pfkA appears to decrease the level of colonization by E. cloacae for seeds that release small quantities of reduced carbon compounds by decreasing the size of the pool of compounds that support rapid growth by this bacterium.
Colonization of the subterranean portions of plants by bacteria can be an essential process when these organisms are used for beneficial purposes, such as plant growth promotion, plant disease control, and bioremediation (2, 3, 5). Several traits, including motility, chemotaxis, salt tolerance, binding to roots, and the production of the O-antigenic side chain of lipopolysaccharide, have been correlated with the colonization of plant surfaces (1, 6, 7, 9, 10, 21). It has also been established that microbial growth is an essential process for colonization (20, 22–26, 28, 29). The complex mixtures of carbohydrates, amino acids, organic acids, and other nutrients (4) released from seeds and roots are thought to support the growth of beneficial bacteria in the spermosphere and rhizosphere. We have used a mutational approach to study the role of the bacterial genes and catabolic pathways and the nutrients supplied by the host plant during growth and colonization of seeds by the potential biocontrol bacterium Enterobacter cloacae 501R3 (17, 18, 22–26).
E. cloacae A-11 is a prototrophic, glycolytic mutant of strain 501R3 with a single mini Tn5-Km transposon insertion in pfkA (22, 26). Roberts et al. found that the populations of strain A-11 on cucumber and radish seeds in natural soil were smaller than the populations of strain 501R3 but the populations of these two strains on pea, soybean, sunflower, and sweet corn seeds were similar (26). It is reasonable to speculate that the mutation in pfkA and the resulting block in glycolysis decrease the populations of E. cloacae A-11 on cucumber and radish seeds by limiting the number of available compounds that support rapid growth by this bacterium. The following findings support this hypothesis. (i) E. cloacae 501R3 was capable of rapid in vitro growth on a large number of carbohydrates found in seed exudates, while strain A-11, which has a mutation in pfkA, had a dramatically lower in vitro growth rate or exhibited no in vitro growth on all of the carbohydrates detected in seed exudates that support rapid growth except fructose (22, 25, 26). Wild-type growth of strain A-11 on fructose was expected since this sugar enters glycolysis after the metabolic block in pfkA (27). And (ii) strains A-11 and 501R3 exhibited comparable levels of colonization of seeds that released relatively large quantities of fructose. Cucumber and radish seeds released substantially less fructose than pea, sunflower, soybean, and sweet corn seeds (26).
We studied the impact of the mutation in pfkA on the growth rate during colonization of seeds by E. cloacae in a sterile sand system by using germinating seeds as continuous sources of reduced carbon compounds. Other methods, such as growth in batch culture or in chemostats containing defined mixtures of reduced carbon compounds, do not approximate spermosphere nutritional conditions (8, 11–13, 16). We used the sterile sand system to demonstrate that the mutation in pfkA decreases the growth rate of E. cloacae during colonization of the spermospheres of certain seeds. The impact of this mutation on the growth rate during colonization by E. cloacae was greatest for the seeds that released the smallest quantities of fructose, other carbohydrates, and amino acids.
(Portions of this work have been published previously [25].)
MATERIALS AND METHODS
Analysis of aqueous seed extracts.
Extracts of cucumber, pea, radish, soybean, sunflower, and sweet corn seeds were prepared essentially as described previously (17, 22, 25) after various incubation periods in sterile distilled water (SDW). The total carbohydrate contents of samples were estimated by using the anthrone assay (15); glucose was used as the standard. Individual carbohydrates were identified and quantified as trifluoracetyl derivatives by using gas chromatography as described previously (25, 31). The total amino acid contents of samples were estimated by using the ninhydrin assay (30); l-leucine was used as the standard.
Estimation of growth rates on seeds.
E. cloacae strains were grown overnight at 35°C and 250 rpm in Luria-Bertani broth (14) supplemented with 100 μg of rifampin per ml for strain 501R3 and 100 μg of rifampin per ml and 50 μg of kanamycin per ml for strain A-11. Overnight cultures were washed, resuspended, and applied to single cucumber (Cucumis sativum cv. Marketmore 76), radish (Raphinus sativus cv. Cherry Bomb), pea (Pisum sativum cv. Sugarsnap), soybean (Glycine max cv. Chesapeake), sunflower (Helianthus giganteus), and sweet corn (Zea mays cv. Stowells Evergreen) seeds, and the seeds were each buried in 4 g of washed, sterile sand containing 4 ml of SDW in a 14-ml sterile snap cap tube; the resulting preparations were incubated at 22°C for 30 h. The seeds were surface disinfested with 5% bleach and washed with SDW before the bacterial suspensions were applied. The numbers of CFU were determined periodically by spiral plating (Spiral Systems, Cincinnati, Ohio) the entire contents of a tube onto Luria-Bertani agar containing 100 μg of cycloheximide per ml and the appropriate antibiotics for each bacterial strain. Experiments were performed with surface-disinfested seeds and sterile sand in order to decrease the effects arising from competition with indigenous microbes for nutrients released from the seeds. In all of the experiments rifampin-resistant, E. cloacae-like colonies represented 90 to 99% of the bacteria detected in the samples.
Nonlinear regression techniques were used to obtain a logistic growth curve for each of the experiments in which we compared growth of strains 501R3 and A-11 on individual plant species. Each experiment was performed with seeds from one plant species and was performed six times with four replicate seeds at each of five sampling times (6, 12, 18, 24, 30 h). The results obtained in all experiments performed with all six plant species were combined prior to analysis. The growth rate and final population size were estimated from each growth curve and were analyzed by using mixed-model analysis of variance techniques (SAS Institute, Cary, N.C.) to determine the fixed effects of the seed type and the bacterial strain and their interactions. Our interpretation of the analyses was based on strain-seed interactions and on pairwise mean comparisons of strains for each seed type.
In vitro growth assays.
E. cloacae 501R3 was grown overnight in M56 salts broth (19) containing 0.2% glycerol at 200 rpm and 32°C. The overnight cultures were centrifuged, washed with 10 mM magnesium sulfate, and suspended in 10 mM magnesium sulfate to an optical density at 540 nm of 1.00. Test tubes containing 5 ml of M56 salts broth supplemented with a carbohydrate at a concentration of 0.2%, an l amino acid at a concentration of 0.5%, or an organic acid at a concentration of 0.2% were inoculated with 100-μl portions of this suspension. The cultures were incubated at 32°C and 200 rpm, and the optical densities at 540 nm were determined periodically. Generation times were calculated as described by Miller (14).
RESULTS AND DISCUSSION
Growth rates on seeds and seed colonization.
Previously, it was shown that glycolysis in E. cloacae A-11 was blocked by a mutation in pfkA and that this gene was most important for colonization of seeds that released limited quantities of reduced carbon compounds. The populations of strain A-11 were significantly smaller than the populations of strain 501R3 on cucumber and radish seeds in sterile sand and in natural soil but not on pea, soybean, sunflower, and sweet corn seeds. Roberts et al. described the strain A-11 phenotype in detail but did not explore the underlying reasons for the smaller strain A-11 populations on cucumber and radish seeds (26).
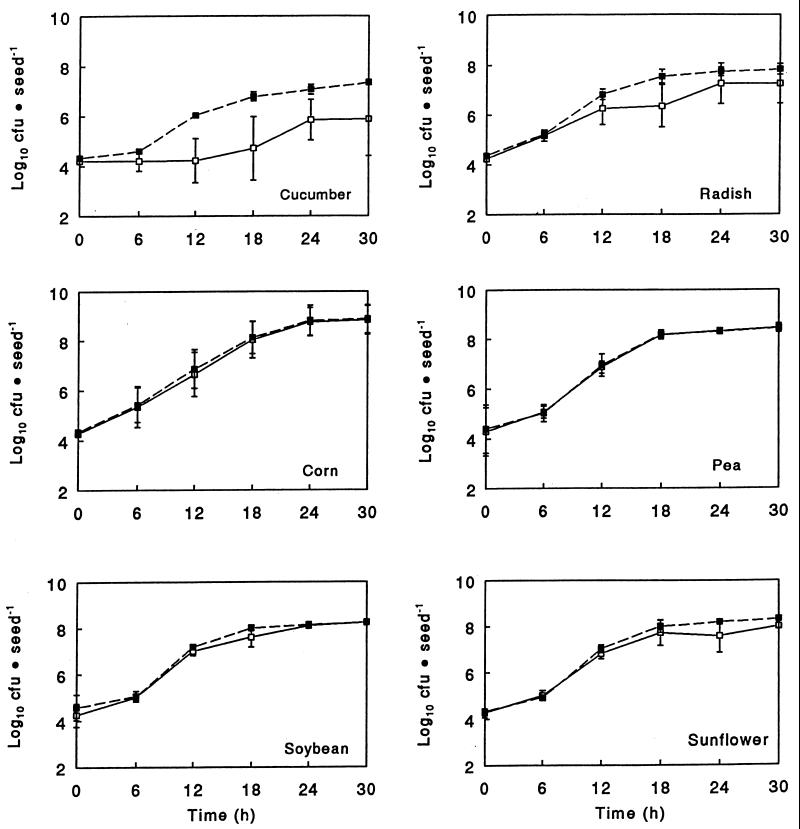
Data obtained in this current study indicated that the lower levels of colonization of cucumber and radish seeds by strain A-11 were due to dramatically lower strain A-11 growth rates on these seeds. A comparison of the growth rates of strains A-11 and 501R3, derived from a logistic model of seed colonization by these strains, showed that the growth rate of strain A-11 was one-half the growth rate of strain 501R3 on radish seeds (significantly different at P ≤ 0.003) and that the strain A-11 growth rate on cucumber seeds was so low that it could not be estimated (Table 1). The growth rate of strain A-11 was only 17% lower on soybean seeds (significantly different at P ≤ 0.001) than the growth rate of strain 501R3 and was similar to the growth rate of strain 501R3 on pea, sunflower, or sweet corn seeds (Table 1). As expected with dramatic reductions in the growth rate, the strain A-11 populations were significantly smaller (P ≤ 0.05) than the strain 501R3 populations on radish seeds 18 h after application and on cucumber seeds 12, 18, 24, and 30 h after application, as determined in these experiments (Fig. 1). The sizes of the strain A-11 and 501R3 populations were similar at all sampling times on pea, soybean, sunflower, and sweet corn seeds, on which the growth rates of strain A-11 were essentially similar to the growth rates of strain 501R3.
TABLE 1.
Growth rates and final population sizes of E. cloacae strains during colonization of various seeds, as estimated with a logistic model
| Seeds | Growth rate (log10 CFU/h)a
|
Final population size (log10 CFU/seed)b
|
||||
|---|---|---|---|---|---|---|
| 501R3 | A-11 | Pr> |T|c | 501R3 | A-11 | Pr> |T|d | |
| Sweet corn | 0.13 | 0.13 | 0.425 | 9.30 | 9.33 | 0.926 |
| Cucumber | 0.16 | NEe | 7.42 | NE | ||
| Pea | 0.20 | 0.19 | 0.206 | 8.53 | 8.58 | 0.237 |
| Radish | 0.20 | 0.10 | 0.003 | 7.87 | 7.96 | 0.738 |
| Soybean | 0.24 | 0.20 | 0.001 | 8.23 | 8.24 | 0.964 |
| Sunflower | 0.22 | 0.23 | 0.845 | 8.35 | 7.94 | 0.140 |
Rate of increase.
The final population sizes were estimated by determining the horizontal asymptotes with the logistic model.
Probability that the growth rates of strains 501R3 and A-11 were not different.
Probability that the final population sizes of strains 501R3 and A-11 were not different.
NE, not estimated (too low to estimate with the logistic model).
FIG. 1.
Population dynamics of E. cloacae A-11 (□) and 501R3 (■) on various types of seeds in sterile sand during the first 30 h after imbibition began. The error bars represent 1 standard deviation from the mean.
In vitro growth on compounds in seed exudates.
The generation times of E. cloacae 501R3 were determined by performing in vitro growth assays in minimal medium containing individual carbohydrates detected in seed exudates. The generation times of strain 501R3 were 69.6 ± 13.1 min on arabinose, 51.4 ± 5.5 min on cellobiose, 60.8 ± 0.1 min on fructose, 58.5 ± 3.1 min on galactose, 51.3 ± 5.9 min on glucose, 70.8 ± 21.2 min on maltose, 55.6 ± 4.5 min on mannitol, 62.3 ± 6.8 min on raffinose, 69.2 ± 1.2 min on ribose, 51.0 ± 2.3 min on sucrose, 55.4 ± 2.2 min on trehalose, and 85.1 ± 11.7 min on xylose. We also determined the generation times of strain 501R3 on individual l amino acids and organic acids that have been reported to be present in seed exudates that support growth of E. cloacae. The generation times of strain 501R3 were 127.8 ± 26.7 min on alanine, 136.7 ± 11.5 min on asparagine, 198.6 ± 30.9 min on proline, 130.8 ± 2.5 min on glutamate, 200.1 ± 51.6 min on glutamine, 376.2 ± 79.8 min on serine, 100.9 ± 7.4 min on pyruvate, and 94.1 ± 2.7 min on malate. The mean generation times during in vitro growth of E. cloacae 501R3 on the carbohydrates, amino acids, and organic acids tested were 61.7 ± 9.9, 195.0 ± 86.5, and 97.5 ± 3.4 min, respectively.
The mutation in pfkA in E. cloacae A-11 reduced the number of carbohydrates in seed exudates that supported wild-type in vitro growth of this bacterium. Of the carbohydrates detected in seed exudates, strain A-11 grew rapidly in vitro only on fructose. The growth of strain A-11 and the growth of strain 501R3 on amino acids and organic acids were similar (26). When we extrapolated from the results of in vitro growth experiments performed with strain 501R3, the net effect of the mutation in pfkA in strain A-11 on growth on compounds released in seed exudates was a shift from rapid growth on carbohydrates to slower growth on amino acids and organic acids.
Seed exudation and growth rate.
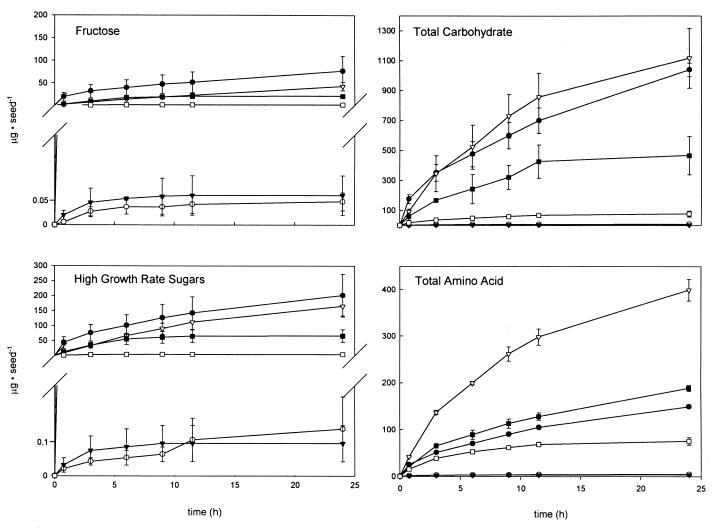
The mutation in pfkA in strain A-11 appears to decrease the growth rate and the level of colonization by E. cloacae on seeds that release small quantities of reduced carbon compounds by limiting the quantity of the compounds that support rapid growth of this bacterium. Three lines of evidence support this hypothesis. First, the decreases in the growth rates of strain A-11 compared to the growth rates of strain 501R3 were substantially greater on cucumber and radish seeds than on the other four types of seeds tested (Table 1). The quantities of compounds released by cucumber and radish seeds that supported rapid growth of E. cloacae were substantially less than the quantities of compounds released by the other four types of seeds tested (Fig. 2). Corn, pea, soybean, and sunflower seeds released 40- to 1,500-fold more carbohydrates (as detected by gas chromatography) that supported rapid in vitro growth of E. cloacae than cucumber or radish seeds released. The carbohydrates that supported rapid growth (defined as in vitro generation times of ≤65 min) were cellobiose, fructose, galactose, glucose, mannitol, raffinose, sucrose, and trehalose. In addition, corn, pea, and soybean seeds released approximately 50- to 100-fold more total carbohydrate than cucumber and radish seeds released during the first 24 h after imbibition began. Sunflower seeds released approximately 10-fold more total carbohydrate than cucumber and radish seeds released (Fig. 2). In general, carbohydrates support much more rapid in vitro growth than amino acids or organic acids support. Second, several sugars were detected, but strain A-11 could grow rapidly only on fructose. Pea, soybean, sunflower, and sweet corn seeds released 56- to 2,500-fold more fructose than cucumber and radish seeds released. In fact, the quantities of fructose released by pea, soybean, and sweet corn seeds during the first 24 h after imbibition began were greater than the combined total quantities of carbohydrates and amino acids released by cucumber and radish seeds. Third, the quantities of sugars available that supported rapid growth of E. cloacae were significantly greater with strain 501R3 than with strain A-11 on cucumber and radish seeds; on these seeds the quantities of sugars that supported rapid growth were very small, the growth rate of strain A-11 was significantly lower than the growth rate of strain 501R3, and the populations of strain A-11 were significantly smaller than the populations of strain 501R3. The concentrations of high-growth-rate sugars available to strains 501R3 and A-11 were 0.13 ± 0.09 and 0.02 ± 0.01 μg per seed, respectively, on cucumber seeds and 0.09 ± 0.05 and 0.03 ± 0.01 μg per seed on radish seeds, respectively.
FIG. 2.
Cumulative quantities of fructose, high-growth-rate sugars, total carbohydrate, and total amino acid released from sweet corn (●), pea (▵), cucumber (○), soybean (■), radish (▴), and sunflower (□) seeds during the first 24 h after imbibition began. The high-growth-rate sugars resulted in in vitro generation times of ≤65 min for strain 501R3. The high-growth-rate sugars are cellobiose, fructose, galactose, glucose, mannitol, raffinose, sucrose, and trehalose. The error bars represent 1 standard deviation from the mean.
The colonization data presented here indicate that the spermospheres of all of the seeds tested were not carbon limited. The estimated final population sizes of strains A-11 and 501R3 were similar despite the inability of strain A-11 to use substantial quantities of reduced carbon that were available to strain 501R3 (Table 1 and Fig. 2). The final population sizes of strain A-11 on cucumber seeds could not be estimated with the model due to the low growth rate. In another study, the population sizes of strains A-11 and 501R3 were similar 96 h after application for all six seed types in seed colonization experiments performed in sterile sand (26). This suggests that required nutrients other than reduced carbon compounds were limiting in the spermospheres of the seeds.
The growth rates of strains A-11 and 501R3 were not strictly correlated with the reduced carbon compounds detected in seed exudates. For example, the estimated growth rate of strain 501R3 on corn seeds was much lower than the growth rate of strain 501R3 on sunflower seeds. Corn seeds release substantially greater quantities of individual sugars that support rapid growth of E. cloacae, total carbohydrates, and total amino acids (Table 1 and Fig. 2). A regression analysis indicated that there was no correlation between the growth rates on seeds of the plant species tested and the quantities of total carbohydrate, total amino acid, fructose, or high-growth-rate sugars released or the rates of release of these compounds (data not shown). High-level correlations between growth rate and exudation of reduced carbon compounds by the seed types tested were not expected under non-carbon-limiting conditions.
Conclusions.
It has been established that microbial growth is an essential process for colonization of plant surfaces (20, 22–26, 28, 29). It can be assumed that the ability of a microorganism to grow on reduced carbon compounds in exudates contributes directly to the ability of the organism to colonize a particular host plant. Hence, the qualitative nature of reduced carbon compounds in exudates with regard to supporting rapid growth should influence colonization. Rapid growth on seeds and roots should be advantageous when it comes to utilizing limiting resources, such as nutrients and space in competitive environments like the plant spermosphere and rhizosphere. It is thought that introduced beneficial bacteria must preempt indigenous bacteria to become established (32).
Studies of the growth of bacteria in batch cultures have shown that different reduced carbon compounds influence microbial growth rates. In these studies bacteria were grown on highly concentrated inorganic minimal media supplemented with reduced carbon compounds at high concentrations (12). However, microbes growing in environmental situations are typically exposed to complex mixtures of substrates which are present at relatively low concentrations (16). Other studies of microbial growth have been performed with low concentrations of defined mixtures of reduced carbon sources in chemostats (8, 11, 13). Unfortunately, the exact qualitative and quantitative compositions of seed exudates and the dynamics of the release of the exudates are not known, which makes extrapolating the results of chemostat studies to the spermosphere and rhizosphere environments difficult.
Studies in which we used E. cloacae 501R3, the near-isogenic strain A-11, seeds, and our sand system provided evidence that directly supports the hypothesis that the ability of a microorganism to rapidly grow on reduced carbon compounds in exudates contributes to its ability to grow on and colonize a particular host plant. Compared with strain A-11, strain 501R3 could grow rapidly on a larger number of compounds present in seed extracts. Strain A-11, with a mutation in pfkA, was not able to grow rapidly on carbohydrates that enter the Embden-Meyeroff-Parnass pathway after fructose 6-phosphate (26, 27). When the quantities of compounds that support rapid growth of E. cloacae were limited, as they are in the cucumber and radish spermospheres, the ability of strain 501R3 to grow on a more diverse collection of sugars resulted in a strain 501R3 growth rate that was dramatically greater than the growth rate of strain A-11. The growth rates of strains A-11 and 501R3 were essentially very similar in pea, soybean, sunflower, and sweet corn spermospheres, in which substantially more sugars that supported rapid growth of both E. cloacae strains were detected. Colonization of cucumber and radish seeds by strain A-11 was reduced, and the growth rate was dramatically affected, while colonization of pea, soybean, sunflower, and sweet corn seeds by strain A-11 was not affected.
Acknowledgments
We thank Kim Brandon and Stanley Tesch for providing excellent technical assistance, Larry Douglas for performing the statistical analysis, and Don Kobayashi and Mark Wilson for reviewing the manuscript.
REFERENCES
- 1.Anderson A J, Habibzadegah-Tari P, Tepper C S. Molecular studies on the role of root surface agglutinin in adherence and colonization by Pseudomonas putida. Appl Environ Microbiol. 1988;54:375–380. doi: 10.1128/aem.54.2.375-380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson T A, Guthrie E A, Walton B T. Bioremediation in the rhizosphere. Environ Sci Technol. 1993;27:2630–2636. [Google Scholar]
- 3.Bull C T, Weller D M, Thomashow L S. Relationship between root colonization and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phytopathology. 1991;81:954–959. [Google Scholar]
- 4.Curl E A, Truelove B. The rhizosphere. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 5.Davison J. Plant beneficial bacteria. Bio/Technology. 1988;6:282–286. [Google Scholar]
- 6.de Weger L A, van der Vlugt C I M, Wijfges A H M, Bakker P A H M, Schippers B, Lugtenberg B. Flagella of a plant-growth-stimulating Pseudomonas fluorescens are required for colonization of potato roots. J Bacteriol. 1987;169:2769–2773. doi: 10.1128/jb.169.6.2769-2773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Weger L A, Bakker P A H M, Schippers B, van Loosdrecht M C M, Lugtenberg B J J. Pseudomonas spp. with mutational changes in the O-antigenic side chain of their lipopolysaccharide and affected in their ability to colonize potato roots. NATO Adv Study Inst Ser H. 1989;36:197–202. [Google Scholar]
- 8.Egli T, Lendenmann U, Snozzi M. Kinetics of microbial growth with mixtures of carbon sources. Antonie Leewenhoek. 1993;63:289–298. doi: 10.1007/BF00871224. [DOI] [PubMed] [Google Scholar]
- 9.Gamliel A, Katan J. Chemotaxis of fluorescent pseudomonads towards seed exudates and germinating seeds in solarized soil. Phytopathology. 1992;82:328–332. [Google Scholar]
- 10.Heinrich D, Hess D. Chemotactic attraction of Azospirillum lipoferum by wheat roots and characterization of some attractants. Can J Microbiol. 1985;31:27–31. [Google Scholar]
- 11.Kovarova K, Kach A, Zehnder A J B, Egli T. Cultivation of Escherichia coli with mixtures of 3-phenylpropionic acid and glucose: steady-state growth kinetics. Appl Environ Microbiol. 1997;63:2619–2624. doi: 10.1128/aem.63.7.2619-2624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lendenmann U, Snozzi M, Egli T. Kinetics of the simultaneous utilization of sugar mixtures by Escherichia coli in continuous culture. Appl Environ Microbiol. 1996;62:1493–1499. doi: 10.1128/aem.62.5.1493-1499.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lendenmann U, Egli T. Is Escherichia coli growing in glucose-limited chemostat culture able to utilize other sugars without lag? Microbiology. 1995;141:71–78. doi: 10.1099/00221287-141-1-71. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 15.Morris D L. Quantitative determination of carbohydrates with Dreywood's anthrone reagent. Science. 1948;107:254–255. doi: 10.1126/science.107.2775.254. [DOI] [PubMed] [Google Scholar]
- 16.Munster U. Concentrations and fluxes of organic carbon substrates in the aquatic environment. Antonie Leeuwenhoek. 1993;63:243–274. doi: 10.1007/BF00871222. [DOI] [PubMed] [Google Scholar]
- 17.Nelson E B, Chao W L, Norton J M, Nash G T, Harman G E. Attachment of Enterobacter cloacae to hyphae of Pythium ultimum: possible role in the biological control of Pythium preemergence damping-off. Phytopathology. 1986;76:327–335. [Google Scholar]
- 18.Nelson E B. Biological control of Pythium seed rot and preemergence damping-off of cotton with Enterobacter cloacae and Erwinia herbicola applied as seed treatments. Plant Dis. 1988;72:140–142. [Google Scholar]
- 19.Nguyen N D, Gottgert M, Singh M, Klingmuller W. Nif-hybrids of Enterobacter cloacae: selection for nif-gene integration with nif-plasmids containing the Mu transposon. Mol Gen Genet. 1983;192:439–443. [Google Scholar]
- 20.O'Sullivan D J, O'Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polonenko D R, Dumbroff E B, Mayfield C I. Microbial responses to salt-induced osmotic stress. II. Population changes in the rhizoplane and rhizosphere. Plant Soil. 1981;63:415–426. [Google Scholar]
- 22.Roberts D P, Sheets C J, Hartung J S. Evidence for proliferation of Enterobacter cloacae on carbohydrates in cucumber and pea spermosphere. Can J Microbiol. 1992;38:1128–1134. [Google Scholar]
- 23.Roberts D P, Marty A M, Dery P D, Yucel I, Hartung J S. Amino acids as reduced carbon sources for Enterobacter cloacae during colonization of the spermospheres of crop plants. Soil Biol Biochem. 1996;28:1015–1020. [Google Scholar]
- 24.Roberts D P, Dery P D, Hartung J S. Peptide utilization and colonization of corn, radish and wheat spermospheres by Enterobacter cloacae. Soil Biol Biochem. 1996;28:1109–1111. [Google Scholar]
- 25.Roberts D P, Dery P D, Yucel I, Buyer J, Holtman M A, Kobayashi D Y. Genetic analysis of the seed colonization mutant Enterobacter cloacae A-11. In: Ogashi A, Kobayashi K, Homma Y, Kodama F, Kondo N, Akino S, editors. Plant growth-promoting rhizobacteria. Present status and future prospects. Proceedings of the Fourth International Workshop on Plant Growth-Promoting Rhizobacteria, Japan-OECD Joint Workshop, Sapporo, Japan, October 5–10, 1997. 1997. pp. 330–332. [Google Scholar]
- 26.Roberts D P, Dery P D, Yucel I, Buyer J, Holtman M A, Kobayashi D Y. Role of pfkA and general carbohydrate catabolism in seed colonization by Enterobacter cloacae. Appl Environ Microbiol. 1999;65:2513–2519. doi: 10.1128/aem.65.6.2513-2519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roehl R A, Vinopal R T. Lack of glucose phosphotransferase function in phosphofructokinase mutants of Escherichia coli. J Bacteriol. 1976;126:852–860. doi: 10.1128/jb.126.2.852-860.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons M, Van Der Bij A J, Brand I, de Weger L A, Wijffelman C A, Lugtenberg B J J. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol Plant-Microbe Interact. 1996;9:600–607. doi: 10.1094/mpmi-9-0600. [DOI] [PubMed] [Google Scholar]
- 29.Simons M, Permentier H P, de Weger L A, Wijffelman C A, Lugtenberg B J J. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant-Microbe Interact. 1997;10:102–106. [Google Scholar]
- 30.Spies J R. Colorimetric procedures for amino acids. In: Colowick S P, Kaplan N O, editors. Methods in enzymology. III. New York, N.Y: Academic Press; 1957. pp. 467–471. [Google Scholar]
- 31.Sullivan J E, Schewe L R. Preparation and gas chromatography of highly volatile trifluoroacetylated carbohydrates using N-methyl bis[trifluoroacetamide] J Chromatogr Sci. 1977;15:196–197. [Google Scholar]
- 32.Weller D M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]